本文主要是介绍LLM生成模型在生物基因DNA应用:HyenaDNA,希望对大家解决编程问题提供一定的参考价值,需要的开发者们随着小编来一起学习吧!
参考:
https://github.com/HazyResearch/hyena-dna
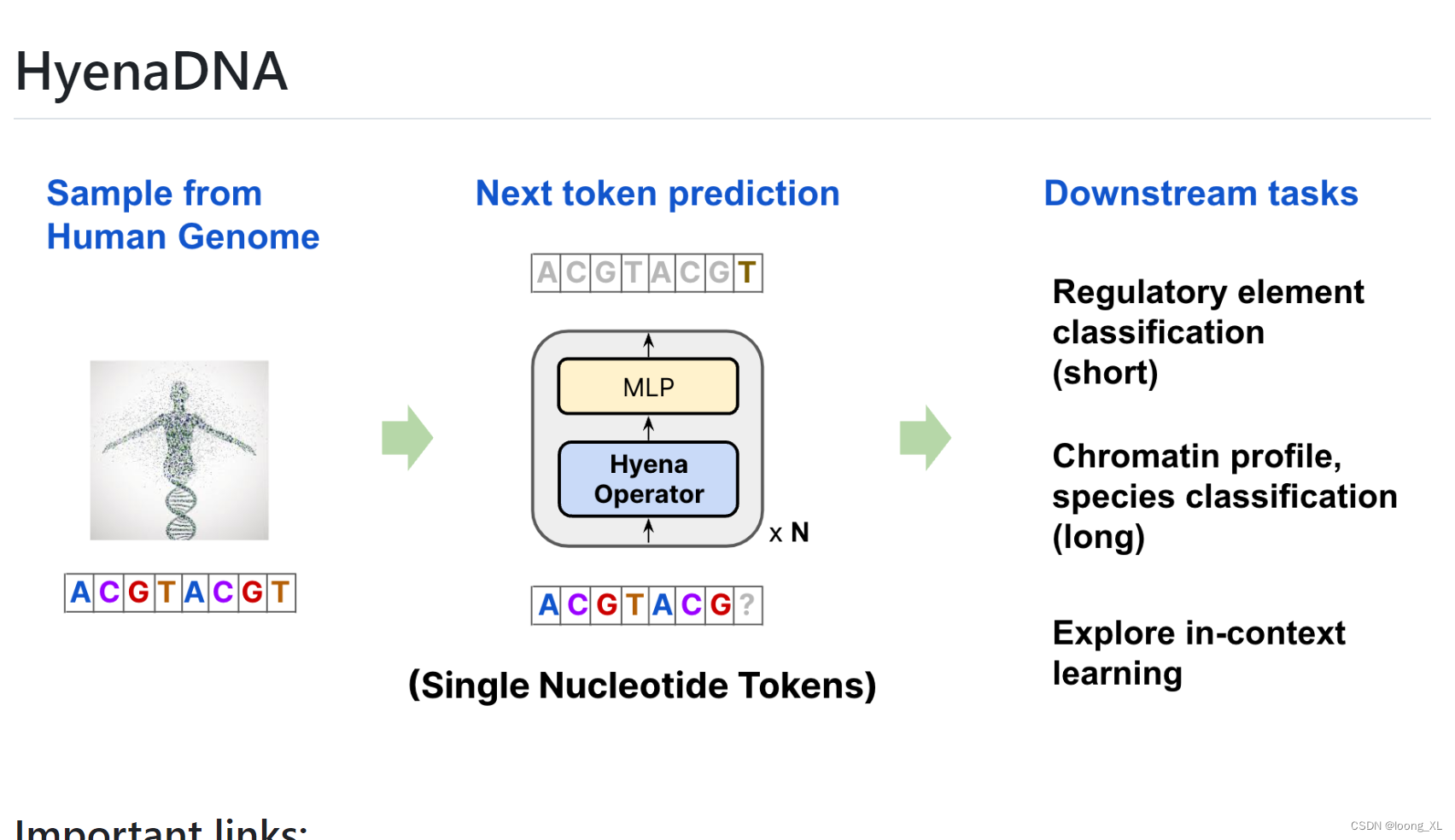
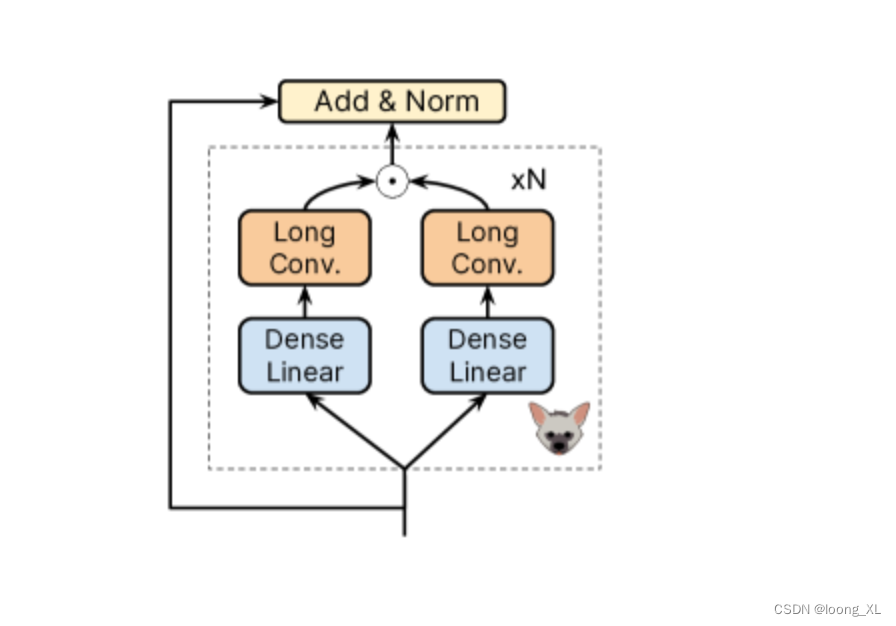
整体框架基本就是GPT模型架构
不一样的就是𝖧𝗒𝖾𝗇𝖺𝖣𝖭𝖠 block ,主要是GPT的多重自注意力层引入了cnn
特征向量提取
# python huggingface.py
#@title Single example
import json
import os
import subprocess
# import transformers
from transformers import PreTrainedModeldef inference_single():'''this selects which backbone to use, and grabs weights/ config from HF4 options:'hyenadna-tiny-1k-seqlen' # fine-tune on colab ok'hyenadna-small-32k-seqlen''hyenadna-medium-160k-seqlen' # inference only on colab'hyenadna-medium-450k-seqlen' # inference only on colab'hyenadna-large-1m-seqlen' # inference only on colab'''# you only need to select which model to use here, we'll do the rest!pretrained_model_name = 'hyenadna-small-32k-seqlen'max_lengths = {'hyenadna-tiny-1k-seqlen': 1024,'hyenadna-small-32k-seqlen': 32768,'hyenadna-medium-160k-seqlen': 160000,'hyenadna-medium-450k-seqlen': 450000, # T4 up to here'hyenadna-large-1m-seqlen': 1_000_000, # only A100 (paid tier)}max_length = max_lengths[pretrained_model_name] # auto selects# data settings:use_padding = Truerc_aug = False # reverse complement augmentationadd_eos = False # add end of sentence token# we need these for the decoder head, if usinguse_head = Falsen_classes = 2 # not used for embeddings only# you can override with your own backbone config here if you want,# otherwise we'll load the HF one in Nonebackbone_cfg = Nonedevice = 'cuda' if torch.cuda.is_available() else 'cpu'print("Using device:", device)# instantiate the model (pretrained here)if pretrained_model_name in ['hyenadna-tiny-1k-seqlen','hyenadna-small-32k-seqlen','hyenadna-medium-160k-seqlen','hyenadna-medium-450k-seqlen','hyenadna-large-1m-seqlen']:# use the pretrained Huggingface wrapper insteadmodel = HyenaDNAPreTrainedModel.from_pretrained('./checkpoints',pretrained_model_name,download=True,config=backbone_cfg,device=device,use_head=use_head,n_classes=n_classes,)# from scratchelif pretrained_model_name is None:model = HyenaDNAModel(**backbone_cfg, use_head=use_head, n_classes=n_classes)# create tokenizertokenizer = CharacterTokenizer(characters=['A', 'C', 'G', 'T', 'N'], # add DNA characters, N is uncertainmodel_max_length=max_length + 2, # to account for special tokens, like EOSadd_special_tokens=False, # we handle special tokens elsewherepadding_side='left', # since HyenaDNA is causal, we pad on the left)#### Single embedding example ##### create a sample 450k long, preparesequence = 'ACTG' * int(max_length/4)tok_seq = tokenizer(sequence)tok_seq = tok_seq["input_ids"] # grab ids# place on device, convert to tensortok_seq = torch.LongTensor(tok_seq).unsqueeze(0) # unsqueeze for batch dimtok_seq = tok_seq.to(device)# prep model and forwardmodel.to(device)model.eval()with torch.inference_mode():embeddings = model(tok_seq)print(embeddings.shape) # embeddings here!# # uncomment to run! (to get embeddings)
inference_single()这篇关于LLM生成模型在生物基因DNA应用:HyenaDNA的文章就介绍到这儿,希望我们推荐的文章对编程师们有所帮助!