本文主要是介绍论文笔记(含代码)Tumor attention networks: Better feature selection, better tumor segmentation,希望对大家解决编程问题提供一定的参考价值,需要的开发者们随着小编来一起学习吧!
肿瘤注意网络:更好的特征选择,更好的肿瘤分割
ELSEVIER Neural Networks 2021
1.提出了一种精确的自动肿瘤分割方法(TA-Net),通过充分利用卷积神经网络和视觉注意机制,用于临床肝脏计算机断层扫描。
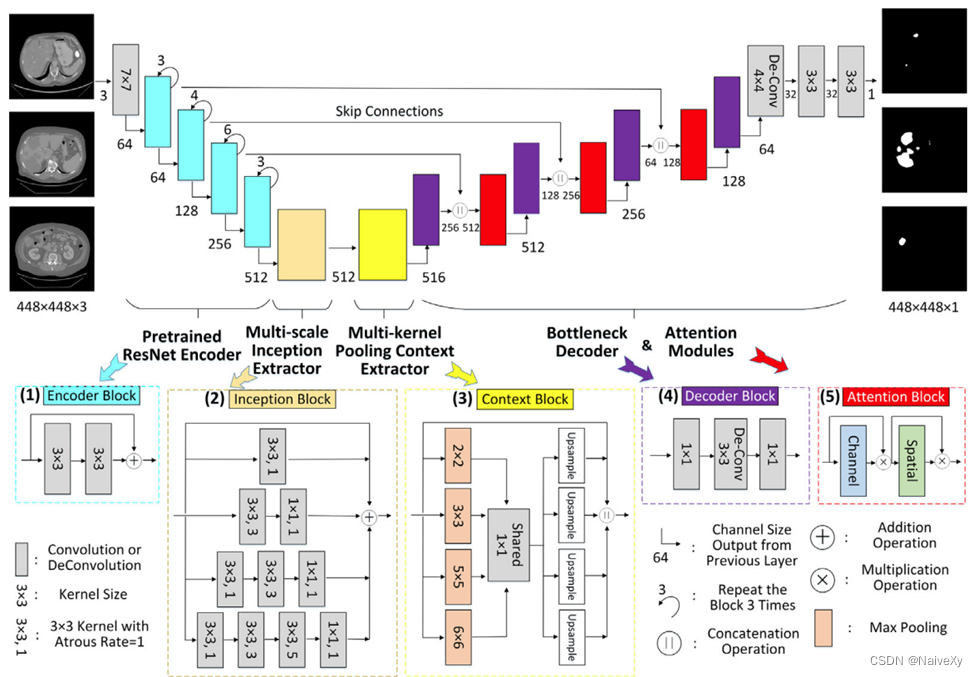
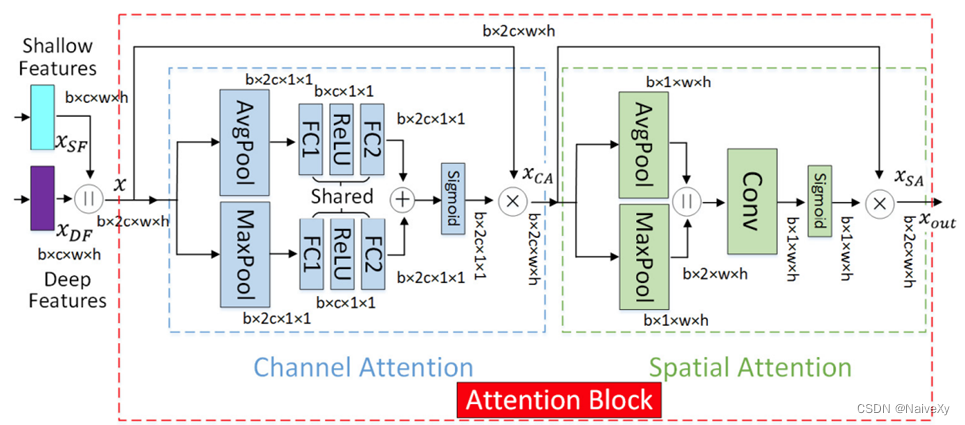
2.设计了一个新的肝脏肿瘤分割管道,从不同角度利用各种类型的网络模块,如Encoder Blocks(预训练网络),各种模块和块重复几次(网络深度)、Inception Blocks 和 Context Blocks(网络宽度)、Decoder Blocks(参数缩减)、Skip Connections(信息融合)和 Tumor Attention Blocks(视觉注意方案和网络基数)。
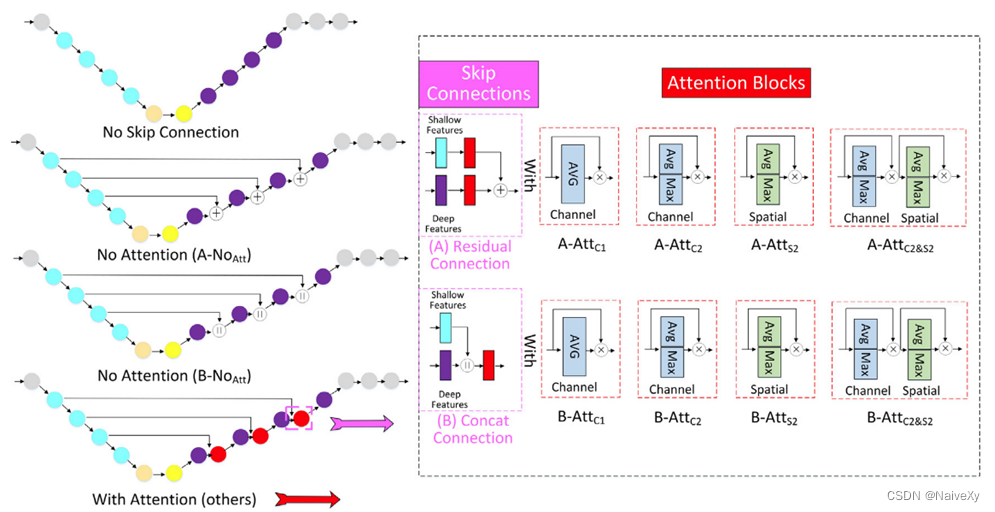
3.对两种流行的skip connection(Residual残差 Connection vs Concat Connection)和无skip connection进行了深入分析和比较:
评估指标
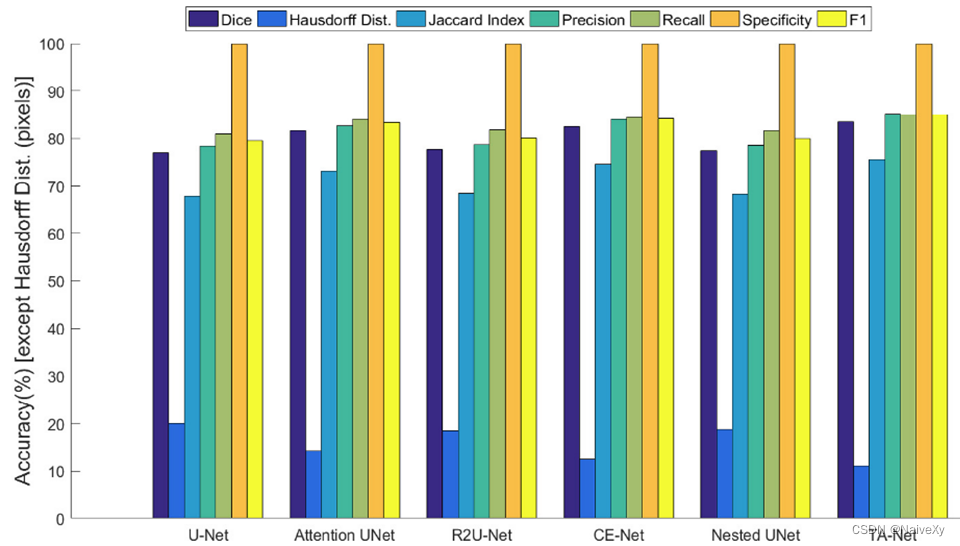
Dice coefficient (DC) Volume Overlap Error (VOE) Relative Volume Error (RVD)
Average Symmetric Surface Distance (ASD) Maximum Surface Distance (MSD)
Dice 系数、体积重叠误差(VOE)、相对体积误差(RVD)
平均对称表面距离(ASD/ASDD)、均方根对称面距离(RMSD)
import torch
import torch.nn as nn
from torchvision import models
import torch.nn.functional as Ffrom functools import partialimport Constantsnonlinearity = partial(F.relu, inplace=True)class DACblock(nn.Module):def __init__(self, channel):super(DACblock, self).__init__()self.dilate1 = nn.Conv2d(channel, channel, kernel_size=3, dilation=1, padding=1)self.dilate2 = nn.Conv2d(channel, channel, kernel_size=3, dilation=3, padding=3)self.dilate3 = nn.Conv2d(channel, channel, kernel_size=3, dilation=5, padding=5)self.conv1x1 = nn.Conv2d(channel, channel, kernel_size=1, dilation=1, padding=0)for m in self.modules():if isinstance(m, nn.Conv2d) or isinstance(m, nn.ConvTranspose2d):if m.bias is not None:m.bias.data.zero_()def forward(self, x):dilate1_out = nonlinearity(self.dilate1(x))dilate2_out = nonlinearity(self.conv1x1(self.dilate2(x)))dilate3_out = nonlinearity(self.conv1x1(self.dilate2(self.dilate1(x))))dilate4_out = nonlinearity(self.conv1x1(self.dilate3(self.dilate2(self.dilate1(x)))))out = x + dilate1_out + dilate2_out + dilate3_out + dilate4_outreturn outclass DACblock_without_atrous(nn.Module):def __init__(self, channel):super(DACblock_without_atrous, self).__init__()self.dilate1 = nn.Conv2d(channel, channel, kernel_size=3, dilation=1, padding=1)self.dilate2 = nn.Conv2d(channel, channel, kernel_size=3, dilation=1, padding=1)self.dilate3 = nn.Conv2d(channel, channel, kernel_size=3, dilation=1, padding=1)self.conv1x1 = nn.Conv2d(channel, channel, kernel_size=1, dilation=1, padding=0)for m in self.modules():if isinstance(m, nn.Conv2d) or isinstance(m, nn.ConvTranspose2d):if m.bias is not None:m.bias.data.zero_()def forward(self, x):dilate1_out = nonlinearity(self.dilate1(x))dilate2_out = nonlinearity(self.conv1x1(self.dilate2(x)))dilate3_out = nonlinearity(self.conv1x1(self.dilate2(self.dilate1(x))))dilate4_out = nonlinearity(self.conv1x1(self.dilate3(self.dilate2(self.dilate1(x)))))out = x + dilate1_out + dilate2_out + dilate3_out + dilate4_outreturn outclass DACblock_with_inception(nn.Module):def __init__(self, channel):super(DACblock_with_inception, self).__init__()self.dilate1 = nn.Conv2d(channel, channel, kernel_size=1, dilation=1, padding=0)self.dilate3 = nn.Conv2d(channel, channel, kernel_size=3, dilation=1, padding=1)self.conv1x1 = nn.Conv2d(2 * channel, channel, kernel_size=1, dilation=1, padding=0)for m in self.modules():if isinstance(m, nn.Conv2d) or isinstance(m, nn.ConvTranspose2d):if m.bias is not None:m.bias.data.zero_()def forward(self, x):dilate1_out = nonlinearity(self.dilate1(x))dilate2_out = nonlinearity(self.dilate3(self.dilate1(x)))dilate_concat = nonlinearity(self.conv1x1(torch.cat([dilate1_out, dilate2_out], 1)))dilate3_out = nonlinearity(self.dilate1(dilate_concat))out = x + dilate3_outreturn outclass DACblock_with_inception_blocks(nn.Module):def __init__(self, channel):super(DACblock_with_inception_blocks, self).__init__()self.conv1x1 = nn.Conv2d(channel, channel, kernel_size=1, dilation=1, padding=0)self.conv3x3 = nn.Conv2d(channel, channel, kernel_size=3, dilation=1, padding=1)self.conv5x5 = nn.Conv2d(channel, channel, kernel_size=5, dilation=1, padding=2)self.pooling = nn.MaxPool2d(kernel_size=3, stride=1, padding=1)for m in self.modules():if isinstance(m, nn.Conv2d) or isinstance(m, nn.ConvTranspose2d):if m.bias is not None:m.bias.data.zero_()def forward(self, x):dilate1_out = nonlinearity(self.conv1x1(x))dilate2_out = nonlinearity(self.conv3x3(self.conv1x1(x)))dilate3_out = nonlinearity(self.conv5x5(self.conv1x1(x)))dilate4_out = self.pooling(x)out = dilate1_out + dilate2_out + dilate3_out + dilate4_outreturn outclass PSPModule(nn.Module):def __init__(self, features, out_features=1024, sizes=(2, 3, 6, 14)):super().__init__()self.stages = []self.stages = nn.ModuleList([self._make_stage(features, size) for size in sizes])self.bottleneck = nn.Conv2d(features * (len(sizes) + 1), out_features, kernel_size=1)self.relu = nn.ReLU()def _make_stage(self, features, size):prior = nn.AdaptiveAvgPool2d(output_size=(size, size))conv = nn.Conv2d(features, features, kernel_size=1, bias=False)return nn.Sequential(prior, conv)def forward(self, feats):h, w = feats.size(2), feats.size(3)priors = [F.upsample(input=stage(feats), size=(h, w), mode='bilinear') for stage in self.stages] + [feats]bottle = self.bottleneck(torch.cat(priors, 1))return self.relu(bottle)class SPPblock(nn.Module):def __init__(self, in_channels):super(SPPblock, self).__init__()self.pool1 = nn.MaxPool2d(kernel_size=[2, 2], stride=2)self.pool2 = nn.MaxPool2d(kernel_size=[3, 3], stride=3)self.pool3 = nn.MaxPool2d(kernel_size=[5, 5], stride=5)self.pool4 = nn.MaxPool2d(kernel_size=[6, 6], stride=6)self.conv = nn.Conv2d(in_channels=in_channels, out_channels=1, kernel_size=1, padding=0)def forward(self, x):self.in_channels, h, w = x.size(1), x.size(2), x.size(3)self.layer1 = F.upsample(self.conv(self.pool1(x)), size=(h, w), mode='bilinear')self.layer2 = F.upsample(self.conv(self.pool2(x)), size=(h, w), mode='bilinear')self.layer3 = F.upsample(self.conv(self.pool3(x)), size=(h, w), mode='bilinear')self.layer4 = F.upsample(self.conv(self.pool4(x)), size=(h, w), mode='bilinear')out = torch.cat([self.layer1, self.layer2, self.layer3, self.layer4, x], 1)return outclass DecoderBlock(nn.Module):def __init__(self, in_channels, n_filters):super(DecoderBlock, self).__init__()self.conv1 = nn.Conv2d(in_channels, in_channels // 4, 1)self.norm1 = nn.BatchNorm2d(in_channels // 4)self.relu1 = nonlinearityself.deconv2 = nn.ConvTranspose2d(in_channels // 4, in_channels // 4, 3, stride=2, padding=1, output_padding=1)self.norm2 = nn.BatchNorm2d(in_channels // 4)self.relu2 = nonlinearityself.conv3 = nn.Conv2d(in_channels // 4, n_filters, 1)self.norm3 = nn.BatchNorm2d(n_filters)self.relu3 = nonlinearitydef forward(self, x):x = self.conv1(x)x = self.norm1(x)x = self.relu1(x)x = self.deconv2(x)x = self.norm2(x)x = self.relu2(x)x = self.conv3(x)x = self.norm3(x)x = self.relu3(x)return xclass ChannelMeanAttention(nn.Module):def __init__(self, num_channels):super(ChannelMeanAttention, self).__init__()num_channels_reduced = num_channels // 2self.fc1 = nn.Linear(num_channels, num_channels_reduced, bias=True)self.fc2 = nn.Linear(num_channels_reduced, num_channels, bias=True)self.relu = nonlinearitydef forward(self, input_tensor):batch_size, num_channels, H, W = input_tensor.size()squeeze_tensor = input_tensor.view(batch_size, num_channels, -1).mean(dim=2)fc_out_1 = self.relu(self.fc1(squeeze_tensor))fc_out_2 = F.sigmoid(self.fc2(fc_out_1))a, b = squeeze_tensor.size()output_tensor = torch.mul(input_tensor, fc_out_2.view(a, b, 1, 1))return output_tensorclass ChannelMeanMaxAttention(nn.Module):def __init__(self, num_channels):super(ChannelMeanMaxAttention, self).__init__()num_channels_reduced = num_channels // 2self.fc1 = nn.Linear(num_channels, num_channels_reduced, bias = True)self.fc2 = nn.Linear(num_channels_reduced, num_channels, bias = True)self.relu = nonlinearitydef forward(self, input_tensor):batch_size, num_channels, H, W = input_tensor.size()squeeze_tensor_mean = input_tensor.view(batch_size, num_channels, -1).mean(dim=2)fc_out_1_mean = self.relu(self.fc1(squeeze_tensor_mean))fc_out_2_mean = self.fc2(fc_out_1_mean)squeeze_tensor_max = input_tensor.view(batch_size, num_channels, -1).max(dim=2)[0]fc_out_1_max = self.relu(self.fc1(squeeze_tensor_max))fc_out_2_max = self.fc2(fc_out_1_max) a, b = squeeze_tensor_mean.size()result = torch.Tensor(a, b)result = torch.add(fc_out_2_mean, fc_out_2_max)fc_out_2 = F.sigmoid(result)output_tensor = torch.mul(input_tensor, fc_out_2.view(a, b, 1, 1))return output_tensorclass SpatialAttention(nn.Module):def __init__(self, kernel_size=7):super(SpatialAttention, self).__init__()padding = 3self.conv1 = nn.Conv2d(2, 1, kernel_size, padding=padding, bias=False)self.sigmoid = nn.Sigmoid()def forward(self, x):input_tensor = xavg_out = torch.mean(x, dim=1, keepdim = True)max_out, _ = torch.max(x, dim=1, keepdim = True)x = torch.cat([avg_out, max_out], dim =1)x = self.conv1(x)return self.sigmoid(x) * input_tensorclass TA_Net_(nn.Module):def __init__(self, num_classes=Constants.BINARY_CLASS, num_channels=3):super(TA_Net_, self).__init__()filters = [64, 128, 256, 512]resnet = models.resnet34(pretrained=True)self.firstconv = resnet.conv1self.firstbn = resnet.bn1self.firstrelu = resnet.reluself.firstmaxpool = resnet.maxpoolself.encoder1 = resnet.layer1self.encoder2 = resnet.layer2self.encoder3 = resnet.layer3self.encoder4 = resnet.layer4self.dblock = DACblock(512)self.spp = SPPblock(512)self.decoder4 = DecoderBlock(516, filters[2])self.channelmeanmaxattention1 = ChannelMeanMaxAttention(filters[2]*2)self.spatialattention1 = SpatialAttention()self.decoder3 = DecoderBlock(filters[2]*2, filters[1])self.channelmeanmaxattention2 = ChannelMeanMaxAttention(filters[1]*2)self.spatialattention2 = SpatialAttention()self.decoder2 = DecoderBlock(filters[1]*2, filters[0])self.channelmeanmaxattention3 = ChannelMeanMaxAttention(filters[0]*2)self.spatialattention3 = SpatialAttention()self.decoder1 = DecoderBlock(filters[0]*2, filters[0])self.finaldeconv1 = nn.ConvTranspose2d(filters[0], 32, 4, 2, 1)self.finalrelu1 = nonlinearityself.finalconv2 = nn.Conv2d(32, 32, 3, padding=1)self.finalrelu2 = nonlinearityself.finalconv3 = nn.Conv2d(32, num_classes, 3, padding=1)def forward(self, x):# Encoderx = self.firstconv(x)x = self.firstbn(x)x = self.firstrelu(x)x = self.firstmaxpool(x)e1 = self.encoder1(x)e2 = self.encoder2(e1)e3 = self.encoder3(e2)e4 = self.encoder4(e3)# Centere4 = self.dblock(e4)e4 = self.spp(e4)# Decoderd4_before = torch.cat([self.decoder4(e4), e3], 1)d4 = self.channelmeanmaxattention1(d4_before)d4 = self.spatialattention1(d4)d3_before = torch.cat([self.decoder3(d4), e2], 1)d3 = self.channelmeanmaxattention2(d3_before)d3 = self.spatialattention2(d3)d2_before = torch.cat([self.decoder2(d3), e1], 1)d2 = self.channelmeanmaxattention3(d2_before)d2 = self.spatialattention3(d2)d1 = self.decoder1(d2)out = self.finaldeconv1(d1)out = self.finalrelu1(out)out = self.finalconv2(out)out = self.finalrelu2(out)out = self.finalconv3(out)return F.sigmoid(out), [d4_before, d3_before, d2_before] # pang for before#return F.sigmoid(out), [d4,d3,d2]def output_features(self, x): # this way doesnot work.# Encoderx = self.firstconv(x)x = self.firstbn(x)x = self.firstrelu(x)x = self.firstmaxpool(x)e1 = self.encoder1(x)e2 = self.encoder2(e1)e3 = self.encoder3(e2)e4 = self.encoder4(e3)# Centere4 = self.dblock(e4)e4 = self.spp(e4)# Decoderd4 = torch.cat([self.decoder4(e4), e3], 1)d4 = self.channelmeanmaxattention1(d4)d4 = self.spatialattention1(d4)d3 = torch.cat([self.decoder3(d4), e2], 1)d3 = self.channelmeanmaxattention2(d3)d3 = self.spatialattention2(d3)d2 = torch.cat([self.decoder2(d3), e1], 1)d2 = self.channelmeanmaxattention3(d2)d2 = self.spatialattention3(d2)d1 = self.decoder1(d2)out = self.finaldeconv1(d1)out = self.finalrelu1(out)out = self.finalconv2(out)out = self.finalrelu2(out)out = self.finalconv3(out)return d4, d3, d2这篇关于论文笔记(含代码)Tumor attention networks: Better feature selection, better tumor segmentation的文章就介绍到这儿,希望我们推荐的文章对编程师们有所帮助!






