本文主要是介绍Titanic: Machine Learning from Disaster,希望对大家解决编程问题提供一定的参考价值,需要的开发者们随着小编来一起学习吧!
A Data Science Framework: To Achieve 99% Accuracy
学习data scientist的思考方式,而不是如何编码。
目录
A Data Science Framework: To Achieve 99% Accuracy
1 怎样处理问题
2 数据科学基本框架
2.1 问题定义
2.2 数据收集
2.3 可用数据准备与数据清洗
2.4 探索性分析
2.5 数据建模
2.6 模型验证及实现
2.7 模型优化及策略
1 怎样处理问题
The goal is to not just learn the whats, but the whys
二元事件结果预测一个经典问题。在外行术语中,这意味着,它要么发生,要么没有发生。例如,你赢了或没有赢,你通过了测试或没有通过,你被接受或没有被接受。常见应用是客户流失或客户保留。另一个流行用例是,医疗卫生的死亡率或生存率分析。二元事件创建了一个有趣的动态,因为我们从统计学上知道,一个随机猜测应该达到50%的准确率,而不需要创建任何算法或编写一行代码。然而,就像自动更正拼写检查技术一样,有时我们人类也会因为太聪明而不能达到自己的目的,实际上不如掷硬币。
2 数据科学基本框架
2.1 问题定义
如果解决方案是数据科学、大数据、机器学习、预测分析、商业智能或任何其他流行词,那么问题是什么?俗话说,不要把马车放在马前面。先问题后需求,先需求后解决,先解决后设计,先设计后技术。在确定我们要解决的实际问题之前,我们常常很快就开始使用新的“闪亮”技术、工具或算法。
项目概要:泰坦尼克号的沉没是历史上最臭名昭著的沉船事件之一。1912年4月15日,泰坦尼克号在处女航中与冰山相撞,2224名乘客和船员中有1502人丧生。这场轰动性的悲剧震惊了国际社会,并导致了更好的船舶安全规则。这次海难造成人员伤亡的原因之一是没有足够的救生艇供乘客和船员使用。尽管在沉船中幸存下来有一些运气因素,但有些人比其他人更可能存活下来,如妇女、儿童和上层阶级。在这个挑战中,我们要求您完成对哪些人可能存活的分析。特别是,我们要求您应用机器学习工具来预测哪些乘客在悲剧中幸存下来。
问题:对所有乘客进行生存分析并预测哪些乘客可能幸存。
2.2 数据收集
JohnNaisbitt在1984年出版的《大趋势》(MegaTrends)一书中写道,我们“沉溺于数据,却坚持求知。”因此,很可能数据集已经以某种形式存在于某个地方。它可能是外部的或内部的,结构化的或非结构化的,静态的或流式的,客观的或主观的,等等。你不必重新发明轮子,你只需要知道在哪里找到它。在下一步中,我们将考虑将“脏数据”转换为“干净数据”。
数据下载:Kaggle's Titanic: Machine Learning from Disaster
2.3 可用数据准备与数据清洗
这一步骤通常被称为data wrangling,是将“野生”数据转变为“可管理”数据的必要过程。data wrangling包括为存储和处理实现数据架构、为质量和控制制定数据治理标准、数据提取(即ETL和Web抓取)和数据清理,以识别异常、丢失或异常的数据点。
2.3.1 导入包
# 基于Python 3的常用包#load packages
import sys #access to system parameters https://docs.python.org/3/library/sys.html
print("Python version: {}". format(sys.version))import pandas as pd # 数据处理和分析建模
print("pandas version: {}". format(pd.__version__))import matplotlib #可视化
print("matplotlib version: {}". format(matplotlib.__version__))import numpy as np #科学计算
print("NumPy version: {}". format(np.__version__))import scipy as sp #科学计算与高级函数集合
print("SciPy version: {}". format(sp.__version__)) import IPython
from IPython import display #pretty printing of dataframes in Jupyter notebook
print("IPython version: {}". format(IPython.__version__)) import sklearn #机器学习算法库
print("scikit-learn version: {}". format(sklearn.__version__))#其他的库
import random
import time#ignore warnings
import warnings
warnings.filterwarnings('ignore')
print('-'*25)# List the files in the data directoryfrom subprocess import check_output
print(check_output(["ls", "../data"]).decode("utf8"))import os #作用同上
print('\n'.join(os.listdir("../data")))
print结果:
Python version: 3.7.3 (default, Mar 27 2019, 17:13:21) [MSC v.1915 64 bit (AMD64)]
pandas version: 0.24.2
matplotlib version: 3.0.3
NumPy version: 1.16.4
SciPy version: 1.2.1
IPython version: 7.4.0
scikit-learn version: 0.21.1
--------------------------------------------------
gender_submission.csv
test.csv
train.csv导入常见的算法包
#Common Model Algorithms
from sklearn import svm, tree, linear_model, neighbors, naive_bayes, ensemble, discriminant_analysis, gaussian_process
from xgboost import XGBClassifier#Common Model Helpers
from sklearn.preprocessing import OneHotEncoder, LabelEncoder
from sklearn import feature_selection
from sklearn import model_selection
from sklearn import metrics#可视化
import matplotlib as mpl
import matplotlib.pyplot as plt
import matplotlib.pylab as pylab
import seaborn as sns
from pandas.tools.plotting import scatter_matrix#设置可视化参数
#%matplotlib inline
mpl.style.use('ggplot')
sns.set_style('white')
pylab.rcParams['figure.figsize'] = 12,82.3.2 初见数据
了解你的数据,它是什么样子的(数据类型和值),是什么让它选中(独立/特征变量),它在生活中的意义是什么(依赖/目标变量)。
Survived variable:是结果或因变量。它是一个二元数据类型,1表示生存,0表示不生存。所有其他变量都是潜在预测变量或独立变量。值得注意的是,预测变量越多不意味着模型越好,只有选择正确变量才能使模型更好。
Passengerid and Ticket variables:是随机的唯一标识符,对结果变量没有影响。因此,它们可能被排除在分析之外。
Pclass variable:是ticket类的顺序数据类型,代表社会经济地位(ses),表示1=上层阶级,2=中产阶级,3=下层阶级。
Name variable:是一个名义数据类型。它可以用于特征工程中,从头衔中得出性别,从姓氏中得出家庭规模,从诸如博士或硕士的头衔中得出社会地位。由于这些变量已经存在,我们将利用它来得到Tilte variable,像master一样,是否会产生影响。
Sex and Embarked variables:是一个名义的数据类型。它们将转换为虚拟变量进行数学计算。
Age and Fare variable:是连续的定量数据类型。
SibSp and Parch variables: SibSp代表船上其兄弟姐妹/配偶的数量,PARCH代表船上其父母/子女的数量。两者都是离散的定量数据类型。这可用于特征工程以构建家族大小,并且仅是变量。
Cabin and SES variables:Cabin(座舱)变量是一个名义数据类型,可用于特征工程中,在事故发生时船上的近似位置和甲板上的SES。但是,由于有许多空值,可供价值不大,因此被排除在分析之外。
#import data from file: https://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_csv.html
data_raw = pd.read_csv('../data/train.csv')#a dataset组成: train, test, and (final) validation 在此作者在后面将train.csv分为train set和test set两部分,用于模型构建与测试,test.csv作为validation set.
data_val = pd.read_csv('../data/test.csv')#创建一个data副本用于后续分析处理
#remember python assignment or equal passes by reference vs values, so we use the copy function: https://stackoverflow.com/questions/46327494/python-pandas-dataframe-copydeep-false-vs-copydeep-true-vs
data1 = data_raw.copy(deep = True)#将train set和test set合并处理
data_cleaner = [data1, data_val]#数据概览
print (data_raw.info()) #查看各字段的信息https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.info.html
print(data_raw.head(5)) #显示前5行数据https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.head.html
print(data_raw.tail(5)) #显示最后5行https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.tail.html
print(data_raw.sample(10)) #随机显示10行https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.sample.html
print(data_raw.columns) #查看列名https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.columns.html
print(data_raw.shape) #查看数据集行列分布,几行几列https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.shape.html
print(data_raw.describe()) #查看数据的大体情况https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.describe.html
data_raw.info结果:
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 891 entries, 0 to 890
Data columns (total 12 columns):
PassengerId 891 non-null int64
Survived 891 non-null int64
Pclass 891 non-null int64
Name 891 non-null object
Sex 891 non-null object
Age 714 non-null float64
SibSp 891 non-null int64
Parch 891 non-null int64
Ticket 891 non-null object
Fare 891 non-null float64
Cabin 204 non-null object
Embarked 889 non-null object
dtypes: float64(2), int64(5), object(5)
memory usage: 83.6+ KB
Nonedata_raw.sample(10)结果:

data_raw.describe()结果:

2.3.3 数据清洗的4个C:纠正(Correcting)、填补(Completing)、创建(Creating)和转换(Converting)
1)纠正异常值,2)完成丢失的信息,3)创建新的分析功能,以及4)将字段转换为正确的计算格式来清理数据
1)纠正:检查数据,没有任何异常或不可接受的数据输入。
我们发现在年龄和票价上可能存在潜在的异常值。但是,由于它们是合理的值,我们将等到完成探索性分析后再确定是否应该排除在数据集中。如果它们肯定是不合理的值,例如年龄=800,那么可以直接剔除。但是,当我们从原始值修改数据时要小心,因为可能需要创建精确的模型。
2)填补:缺少值可能很糟糕,因为有些算法不能处理空值。而其他的,如决策树,可以处理空值。因此,在开始建模之前填补空值是很重要的,因为我们将比较和对比几个模型。有两种常见的方法,要么删除记录,要么使用合理的输入填充缺少的值。不建议删除记录,尤其是大部分记录,除非它确实代表一个不完整的记录。相反,最好将缺失的值归集。定性数据的一种基本方法是使用模式输入。定量数据的基本方法是使用平均值、中值或平均值+随机标准差进行输入。
年龄、船舱和SES字段中存在空值。中间方法是根据具体标准使用基本方法,如按平均年龄或按票价和SES上船港口。然而,在部署之前,有更复杂的方法论,应该将其与基础模型进行比较,以确定复杂性是否真的有价值。对于这个数据集,年龄将由中间值输入,座舱属性将被删除,登船将由模式输入。随后的模型迭代可能会修改这个决定,以确定它是否提高了模型的准确性。
3)创建:特征工程就是我们使用现有的特征来创建新的特征,以确定它们是否提供新的信息来预测结果。对于这个数据集,我们将创建一个Title变量来确定它是否在生存中起到作用。
4)转换:最后,我们将数据处理格式化。没有日期或货币格式,只有数据类型格式。我们的分类数据作为对象导入,这使得数学计算变得困难。对于这个数据集,我们将把对象数据类型转换为分类虚拟变量。
print('Train columns with null values:\n', data1.isnull().sum())
print("-"*10)print('Test/Validation columns with null values:\n', data_val.isnull().sum())
print("-"*10)
结果:
Train columns with null values:PassengerId 0
Survived 0
Pclass 0
Name 0
Sex 0
Age 177
SibSp 0
Parch 0
Ticket 0
Fare 0
Cabin 687
Embarked 2
dtype: int64
----------
Test/Validation columns with null values:PassengerId 0
Pclass 0
Name 0
Sex 0
Age 86
SibSp 0
Parch 0
Ticket 0
Fare 1
Cabin 327
Embarked 0
dtype: int64
----------Now that we know what to clean, let's execute our code.
a 数据清洗
Developer Documentation:
- pandas.DataFrame
- pandas.DataFrame.info
- pandas.DataFrame.describe
- Indexing and Selecting Data
- pandas.isnull
- pandas.DataFrame.sum
- pandas.DataFrame.mode
- pandas.DataFrame.copy
- pandas.DataFrame.fillna
- pandas.DataFrame.drop
- pandas.Series.value_counts
- pandas.DataFrame.loc
###填充或删除缺失值
for dataset in data_cleaner: #complete missing age with mediandataset['Age'].fillna(dataset['Age'].median(), inplace = True)#complete embarked with modedataset['Embarked'].fillna(dataset['Embarked'].mode()[0], inplace = True)#complete missing fare with mediandataset['Fare'].fillna(dataset['Fare'].median(), inplace = True)#删除 Cabin feature/column
drop_column = ['PassengerId','Cabin', 'Ticket']
data1.drop(drop_column, axis=1, inplace = True)print(data1.isnull().sum())
print("-"*10)
print(data_val.isnull().sum())结果:
Survived 0
Pclass 0
Name 0
Sex 0
Age 0
SibSp 0
Parch 0
Fare 0
Embarked 0
dtype: int64
----------
PassengerId 0
Pclass 0
Name 0
Sex 0
Age 0
SibSp 0
Parch 0
Ticket 0
Fare 0
Cabin 327
Embarked 0
dtype: int64
###3)创建特征
for dataset in data_cleaner: #Discrete variablesdataset['FamilySize'] = dataset ['SibSp'] + dataset['Parch'] + 1dataset['IsAlone'] = 1 #initialize to yes/1 is alonedataset['IsAlone'].loc[dataset['FamilySize'] > 1] = 0 # now update to no/0 if family size is greater than 1#从name中提取Title: http://www.pythonforbeginners.com/dictionary/python-splitdataset['Title'] = dataset['Name'].str.split(", ", expand=True)[1].str.split(".", expand=True)[0]#Fare与Age连续区间划分; qcut vs cut: https://stackoverflow.com/questions/30211923/what-is-the-difference-between-pandas-qcut-and-pandas-cut#Fare Bins/Buckets using qcut or frequency bins: https://pandas.pydata.org/pandas-docs/stable/generated/pandas.qcut.htmldataset['FareBin'] = pd.qcut(dataset['Fare'], 4)#Age Bins/Buckets using cut or value bins: https://pandas.pydata.org/pandas-docs/stable/generated/pandas.cut.htmldataset['AgeBin'] = pd.cut(dataset['Age'].astype(int), 5)#清除数目较少的Title
print(data1['Title'].value_counts())
stat_min = 10 #公共最小值http://nicholasjjackson.com/2012/03/08/sample-size-is-10-a-magic-number/
title_names = (data1['Title'].value_counts() < stat_min) #this will create a true false series with title name as index#apply and lambda functions 查找与替换: https://community.modeanalytics.com/python/tutorial/pandas-groupby-and-python-lambda-functions/
data1['Title'] = data1['Title'].apply(lambda x: 'Misc' if title_names.loc[x] == True else x)
print(data1['Title'].value_counts())
print("-"*10)#preview data again
data1.info()
data_val.info()
data1.sample(10)结果:
Mr 517
Miss 182
Mrs 125
Master 40
Misc 27
Name: Title, dtype: int64
----------
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 891 entries, 0 to 890
Data columns (total 14 columns):
Survived 891 non-null int64
Pclass 891 non-null int64
Name 891 non-null object
Sex 891 non-null object
Age 891 non-null float64
SibSp 891 non-null int64
Parch 891 non-null int64
Fare 891 non-null float64
Embarked 891 non-null object
FamilySize 891 non-null int64
IsAlone 891 non-null int64
Title 891 non-null object
FareBin 891 non-null category
AgeBin 891 non-null category
dtypes: category(2), float64(2), int64(6), object(4)
memory usage: 85.5+ KB
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 418 entries, 0 to 417
Data columns (total 16 columns):
PassengerId 418 non-null int64
Pclass 418 non-null int64
Name 418 non-null object
Sex 418 non-null object
Age 418 non-null float64
SibSp 418 non-null int64
Parch 418 non-null int64
Ticket 418 non-null object
Fare 418 non-null float64
Cabin 91 non-null object
Embarked 418 non-null object
FamilySize 418 non-null int64
IsAlone 418 non-null int64
Title 418 non-null object
FareBin 418 non-null category
AgeBin 418 non-null category
dtypes: category(2), float64(2), int64(6), object(6)
memory usage: 46.8+ KBdata1.sample(10):
b 格式转换
Developer Documentation:
- Categorical Encoding
- Sklearn LabelEncoder
- Sklearn OneHotEncoder
- Pandas Categorical dtype
- pandas.get_dummies
#转换:Label Encoderlabel = LabelEncoder()
for dataset in data_cleaner: dataset['Sex_Code'] = label.fit_transform(dataset['Sex'])dataset['Embarked_Code'] = label.fit_transform(dataset['Embarked'])dataset['Title_Code'] = label.fit_transform(dataset['Title'])dataset['AgeBin_Code'] = label.fit_transform(dataset['AgeBin'])dataset['FareBin_Code'] = label.fit_transform(dataset['FareBin'])#define target
Target = ['Survived']#define x variables
data1_x = ['Sex','Pclass', 'Embarked', 'Title','SibSp', 'Parch', 'Age', 'Fare', 'FamilySize', 'IsAlone']
data1_x_calc = ['Sex_Code','Pclass', 'Embarked_Code', 'Title_Code','SibSp', 'Parch', 'Age', 'Fare']
data1_xy = Target + data1_x
print('Original X Y: ', data1_xy, '\n')#连续型变量变换
data1_x_bin = ['Sex_Code','Pclass', 'Embarked_Code', 'Title_Code', 'FamilySize', 'AgeBin_Code', 'FareBin_Code']
data1_xy_bin = Target + data1_x_bin
print('Bin X Y: ', data1_xy_bin, '\n')#虚拟变量
data1_dummy = pd.get_dummies(data1[data1_x])
data1_x_dummy = data1_dummy.columns.tolist()
data1_xy_dummy = Target + data1_x_dummy
print('Dummy X Y: ', data1_xy_dummy, '\n')data1_dummy.head()结果:
Original X Y: ['Survived', 'Sex', 'Pclass', 'Embarked', 'Title', 'SibSp', 'Parch', 'Age', 'Fare', 'FamilySize', 'IsAlone'] Bin X Y: ['Survived', 'Sex_Code', 'Pclass', 'Embarked_Code', 'Title_Code', 'FamilySize', 'AgeBin_Code', 'FareBin_Code'] Dummy X Y: ['Survived', 'Pclass', 'SibSp', 'Parch', 'Age', 'Fare', 'FamilySize', 'IsAlone', 'Sex_female', 'Sex_male', 'Embarked_C', 'Embarked_Q', 'Embarked_S', 'Title_Master', 'Title_Misc', 'Title_Miss', 'Title_Mr', 'Title_Mrs']
2.3.4 再次检查
print('Train columns with null values: \n', data1.isnull().sum())
print("-"*10)
print (data1.info())
print("-"*10)print('Test/Validation columns with null values: \n', data_val.isnull().sum())
print("-"*10)
print (data_val.info())
print("-"*10)data_raw.describe(include = 'all')结果:
Train columns with null values: Survived 0
Pclass 0
Name 0
Sex 0
Age 0
SibSp 0
Parch 0
Fare 0
Embarked 0
FamilySize 0
IsAlone 0
Title 0
FareBin 0
AgeBin 0
Sex_Code 0
Embarked_Code 0
Title_Code 0
AgeBin_Code 0
FareBin_Code 0
dtype: int64
----------
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 891 entries, 0 to 890
Data columns (total 19 columns):
Survived 891 non-null int64
Pclass 891 non-null int64
Name 891 non-null object
Sex 891 non-null object
Age 891 non-null float64
SibSp 891 non-null int64
Parch 891 non-null int64
Fare 891 non-null float64
Embarked 891 non-null object
FamilySize 891 non-null int64
IsAlone 891 non-null int64
Title 891 non-null object
FareBin 891 non-null category
AgeBin 891 non-null category
Sex_Code 891 non-null int64
Embarked_Code 891 non-null int64
Title_Code 891 non-null int64
AgeBin_Code 891 non-null int64
FareBin_Code 891 non-null int64
dtypes: category(2), float64(2), int64(11), object(4)
memory usage: 120.3+ KB
None
----------
Test/Validation columns with null values: PassengerId 0
Pclass 0
Name 0
Sex 0
Age 0
SibSp 0
Parch 0
Ticket 0
Fare 0
Cabin 327
Embarked 0
FamilySize 0
IsAlone 0
Title 0
FareBin 0
AgeBin 0
Sex_Code 0
Embarked_Code 0
Title_Code 0
AgeBin_Code 0
FareBin_Code 0
dtype: int64
----------
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 418 entries, 0 to 417
Data columns (total 21 columns):
PassengerId 418 non-null int64
Pclass 418 non-null int64
Name 418 non-null object
Sex 418 non-null object
Age 418 non-null float64
SibSp 418 non-null int64
Parch 418 non-null int64
Ticket 418 non-null object
Fare 418 non-null float64
Cabin 91 non-null object
Embarked 418 non-null object
FamilySize 418 non-null int64
IsAlone 418 non-null int64
Title 418 non-null object
FareBin 418 non-null category
AgeBin 418 non-null category
Sex_Code 418 non-null int64
Embarked_Code 418 non-null int64
Title_Code 418 non-null int64
AgeBin_Code 418 non-null int64
FareBin_Code 418 non-null int64
dtypes: category(2), float64(2), int64(11), object(6)
memory usage: 63.1+ KB
None
----------data_raw.describe(include = 'all')结果:
2.3.5 数据集划分
如前所述,所提供的测试文件实际上是提交竞赛的验证数据。因此,我们将使用sklearn的train_test_split函数将训练数据拆分为两个数据集:75/25拆分,保证我们的模型不会过拟合。也就是说,该算法特定于一个给定的子集,它不能归纳另一个子集。重要的是,我们的算法没有用过我们用来测试的子集,所以它不会产生“欺骗性”记忆。在后面的部分中,我们还将使用sklearn的交叉验证功能,该功能将我们的数据集拆分为用于数据建模比较的训练和测试。
#random_state -> seed or control random number generator: https://www.quora.com/What-is-seed-in-random-number-generation
train1_x, test1_x, train1_y, test1_y = model_selection.train_test_split(data1[data1_x_calc], data1[Target], random_state = 0)
train1_x_bin, test1_x_bin, train1_y_bin, test1_y_bin = model_selection.train_test_split(data1[data1_x_bin], data1[Target] , random_state = 0)
train1_x_dummy, test1_x_dummy, train1_y_dummy, test1_y_dummy = model_selection.train_test_split(data1_dummy[data1_x_dummy], data1[Target], random_state = 0)print("Data1 Shape: {}".format(data1.shape))
print("Train1 Shape: {}".format(train1_x.shape))
print("Test1 Shape: {}".format(test1_x.shape))train1_x_bin.head()结果:
Data1 Shape: (891, 19)
Train1 Shape: (668, 8)
Test1 Shape: (223, 8)train_x_bin.head() 结果:
2.4 探索性分析
任何处理过数据的人都知道,垃圾输入,垃圾输出(GIGO)。因此,部署描述性统计和图形统计以在数据集中查找潜在的问题、模式、分类、相关性和比较是很重要的。此外,数据分类(即定性与定量)对于理解和选择正确的假设检验或数据模型也很重要。
在经过之前的操作之后得到干净数据,我们将使用描述性和图形统计来探索我们的数据,以描述和总结我们的变量。在这个阶段,将对特性进行分类,并确定它们与目标变量以及彼此之间的相关性。
#群体生存率的相关分析: https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.groupby.html
for x in data1_x:if data1[x].dtype != 'float64' :print('Survival Correlation by:', x)print(data1[[x, Target[0]]].groupby(x, as_index=False).mean())print('-'*10, '\n')#crosstabs卡方检验: https://pandas.pydata.org/pandas-docs/stable/generated/pandas.crosstab.html
print(pd.crosstab(data1['Title'],data1[Target[0]]))结果:
Survival Correlation by: SexSex Survived
0 female 0.742038
1 male 0.188908
---------- Survival Correlation by: PclassPclass Survived
0 1 0.629630
1 2 0.472826
2 3 0.242363
---------- Survival Correlation by: EmbarkedEmbarked Survived
0 C 0.553571
1 Q 0.389610
2 S 0.339009
---------- Survival Correlation by: TitleTitle Survived
0 Master 0.575000
1 Misc 0.444444
2 Miss 0.697802
3 Mr 0.156673
4 Mrs 0.792000
---------- Survival Correlation by: SibSpSibSp Survived
0 0 0.345395
1 1 0.535885
2 2 0.464286
3 3 0.250000
4 4 0.166667
5 5 0.000000
6 8 0.000000
---------- Survival Correlation by: ParchParch Survived
0 0 0.343658
1 1 0.550847
2 2 0.500000
3 3 0.600000
4 4 0.000000
5 5 0.200000
6 6 0.000000
---------- Survival Correlation by: FamilySizeFamilySize Survived
0 1 0.303538
1 2 0.552795
2 3 0.578431
3 4 0.724138
4 5 0.200000
5 6 0.136364
6 7 0.333333
7 8 0.000000
8 11 0.000000
---------- Survival Correlation by: IsAloneIsAlone Survived
0 0 0.505650
1 1 0.303538
---------- Survived 0 1
Title
Master 17 23
Misc 15 12
Miss 55 127
Mr 436 81
Mrs 26 99定量数据matplotlib可视化:
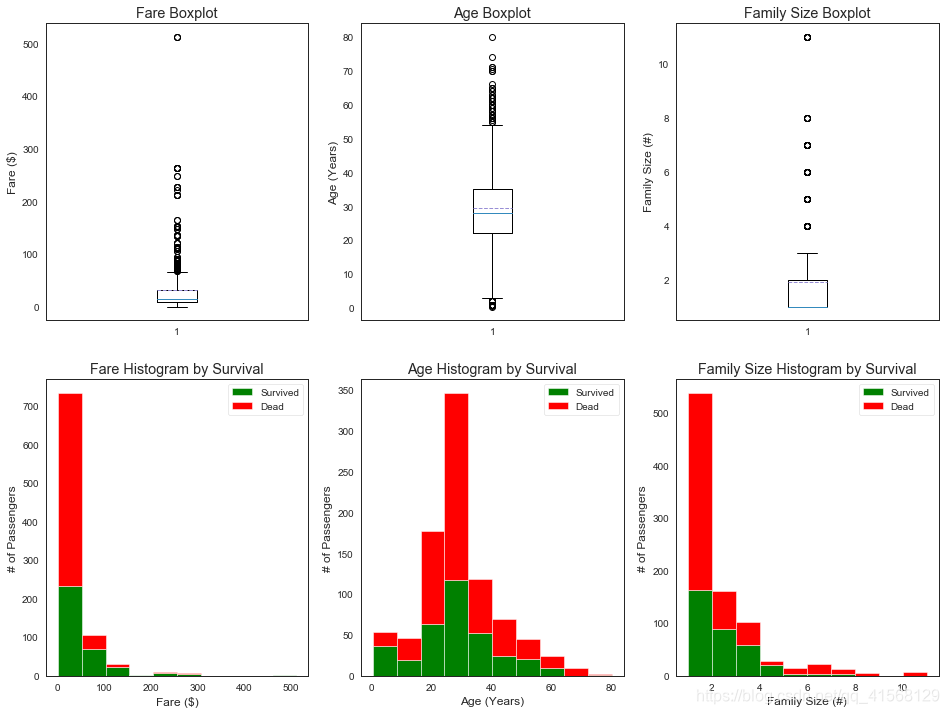
#optional plotting w/pandas: https://pandas.pydata.org/pandas-docs/stable/visualization.html#we will use matplotlib.pyplot: https://matplotlib.org/api/pyplot_api.html to organize our graphics will use figure: https://matplotlib.org/api/_as_gen/matplotlib.pyplot.figure.html#matplotlib.pyplot.figure
#subplot:https://matplotlib.org/api/_as_gen/matplotlib.pyplot.subplot.html#matplotlib.pyplot.subplot
#and subplotS: https://matplotlib.org/api/_as_gen/matplotlib.pyplot.subplots.html?highlight=matplotlib%20pyplot%20subplots#matplotlib.pyplot.subplots#定量数据
plt.figure(figsize=[16,12])plt.subplot(231)
plt.boxplot(x=data1['Fare'], showmeans = True, meanline = True)
plt.title('Fare Boxplot')
plt.ylabel('Fare ($)')plt.subplot(232)
plt.boxplot(data1['Age'], showmeans = True, meanline = True)
plt.title('Age Boxplot')
plt.ylabel('Age (Years)')plt.subplot(233)
plt.boxplot(data1['FamilySize'], showmeans = True, meanline = True)
plt.title('Family Size Boxplot')
plt.ylabel('Family Size (#)')plt.subplot(234)
plt.hist(x = [data1[data1['Survived']==1]['Fare'], data1[data1['Survived']==0]['Fare']], stacked=True, color = ['g','r'],label = ['Survived','Dead'])
plt.title('Fare Histogram by Survival')
plt.xlabel('Fare ($)')
plt.ylabel('# of Passengers')
plt.legend()plt.subplot(235)
plt.hist(x = [data1[data1['Survived']==1]['Age'], data1[data1['Survived']==0]['Age']], stacked=True, color = ['g','r'],label = ['Survived','Dead'])
plt.title('Age Histogram by Survival')
plt.xlabel('Age (Years)')
plt.ylabel('# of Passengers')
plt.legend()plt.subplot(236)
plt.hist(x = [data1[data1['Survived']==1]['FamilySize'], data1[data1['Survived']==0]['FamilySize']], stacked=True, color = ['g','r'],label = ['Survived','Dead'])
plt.title('Family Size Histogram by Survival')
plt.xlabel('Family Size (#)')
plt.ylabel('# of Passengers')
plt.legend()结果:
Seaborn作图:
#使用Seaborn图形进行多变量比较: https://seaborn.pydata.org/api.html#个体特征图
fig, saxis = plt.subplots(2, 3,figsize=(16,12))sns.barplot(x = 'Embarked', y = 'Survived', data=data1, ax = saxis[0,0])
sns.barplot(x = 'Pclass', y = 'Survived', order=[1,2,3], data=data1, ax = saxis[0,1])
sns.barplot(x = 'IsAlone', y = 'Survived', order=[1,0], data=data1, ax = saxis[0,2])sns.pointplot(x = 'FareBin', y = 'Survived', data=data1, ax = saxis[1,0])
sns.pointplot(x = 'AgeBin', y = 'Survived', data=data1, ax = saxis[1,1])
sns.pointplot(x = 'FamilySize', y = 'Survived', data=data1, ax = saxis[1,2])结果:
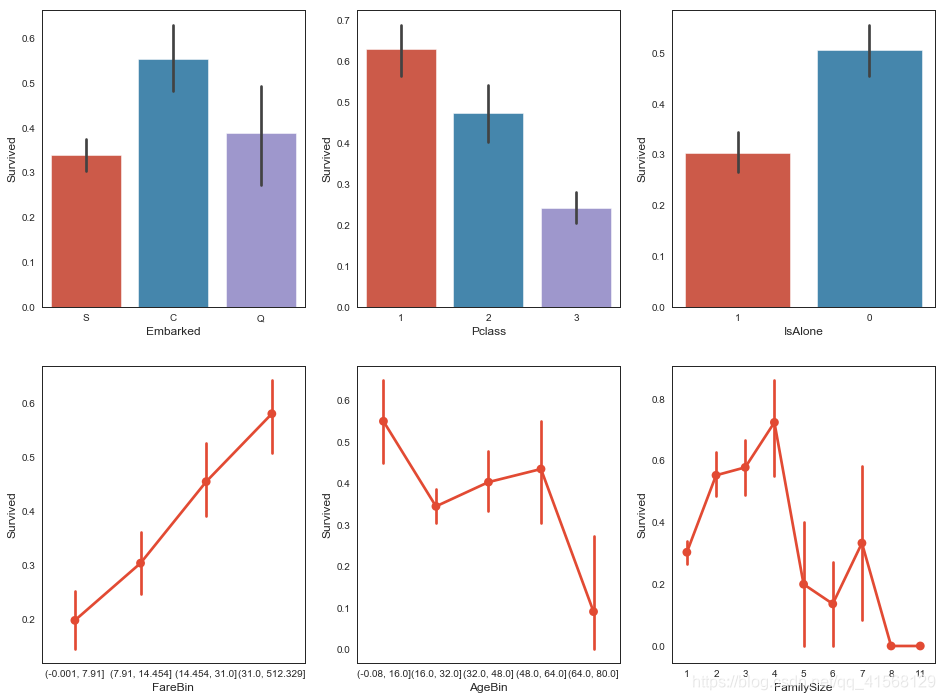
定性变量:
#定性变量: Sex
#we know sex mattered in survival, now let's compare sex and a 2nd feature
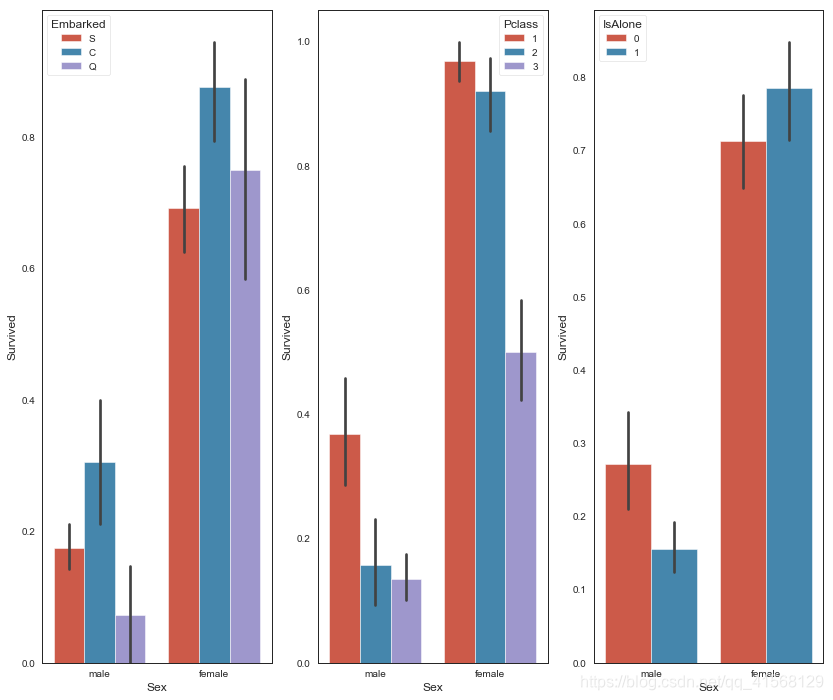
fig, qaxis = plt.subplots(1,3,figsize=(14,12))sns.barplot(x = 'Sex', y = 'Survived', hue = 'Embarked', data=data1, ax = qaxis[0])
axis1.set_title('Sex vs Embarked Survival Comparison')sns.barplot(x = 'Sex', y = 'Survived', hue = 'Pclass', data=data1, ax = qaxis[1])
axis1.set_title('Sex vs Pclass Survival Comparison')sns.barplot(x = 'Sex', y = 'Survived', hue = 'IsAlone', data=data1, ax = qaxis[2])
axis1.set_title('Sex vs IsAlone Survival Comparison')结果:
side-by-side comparisons:
#more side-by-side comparisons
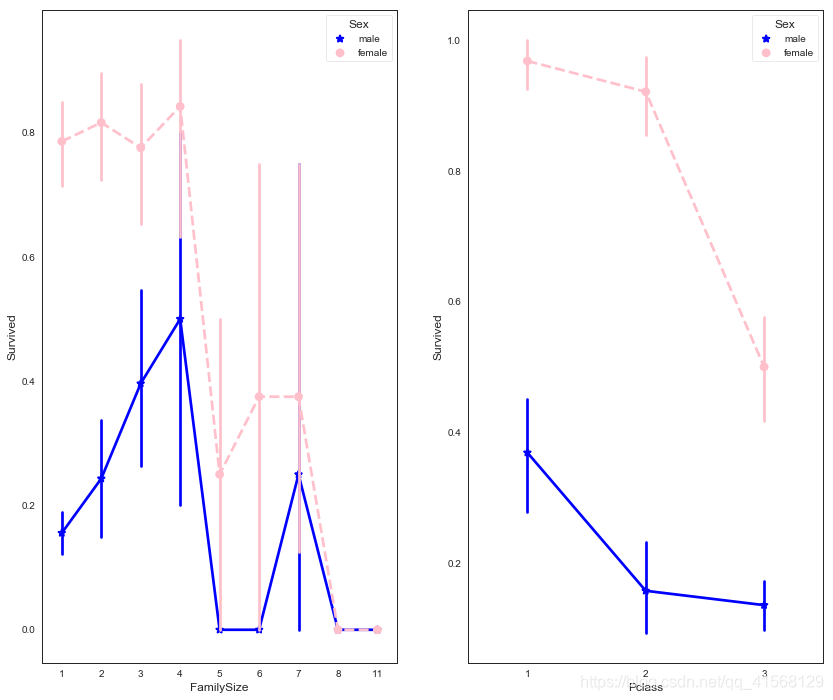
fig, (maxis1, maxis2) = plt.subplots(1, 2,figsize=(14,12))#how does family size factor with sex & survival compare
sns.pointplot(x="FamilySize", y="Survived", hue="Sex", data=data1,palette={"male": "blue", "female": "pink"},markers=["*", "o"], linestyles=["-", "--"], ax = maxis1)#how does class factor with sex & survival compare
sns.pointplot(x="Pclass", y="Survived", hue="Sex", data=data1,palette={"male": "blue", "female": "pink"},markers=["*", "o"], linestyles=["-", "--"], ax = maxis2)结果:
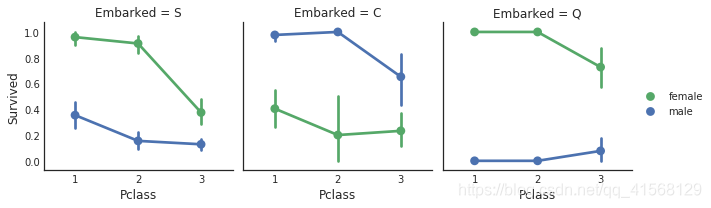
#how does embark port factor with class, sex, and survival compare facetgrid: https://seaborn.pydata.org/generated/seaborn.FacetGrid.html
e = sns.FacetGrid(data1, col = 'Embarked')
e.map(sns.pointplot, 'Pclass', 'Survived', 'Sex', ci=95.0, palette = 'deep')
e.add_legend()结果:
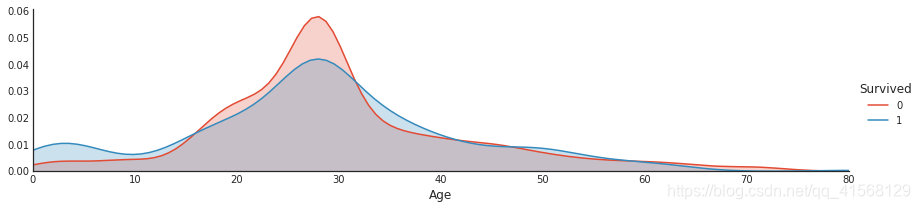
#plot distributions of age of passengers who survived or did not survive
a = sns.FacetGrid( data1, hue = 'Survived', aspect=4 )
a.map(sns.kdeplot, 'Age', shade= True )
a.set(xlim=(0 , data1['Age'].max()))
a.add_legend()结果:
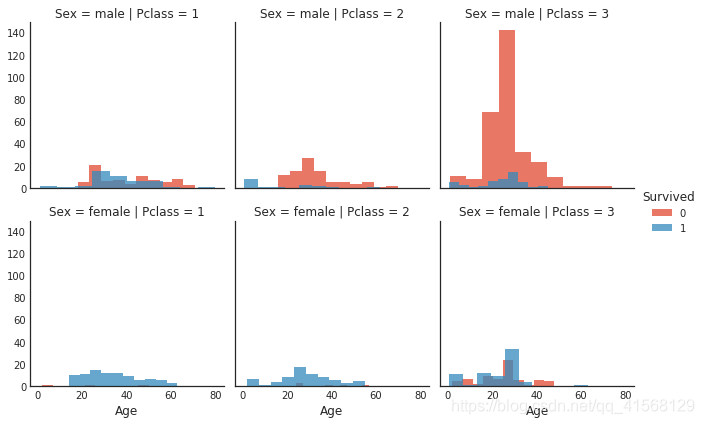
#histogram comparison of sex, class, and age by survival
h = sns.FacetGrid(data1, row = 'Sex', col = 'Pclass', hue = 'Survived')
h.map(plt.hist, 'Age', alpha = .75)
h.add_legend()结果:
#两两比对
pp = sns.pairplot(data1, hue = 'Survived', palette = 'deep', size=1.2, diag_kind = 'kde', diag_kws=dict(shade=True), plot_kws=dict(s=10) )
pp.set(xticklabels=[])结果:

#correlation heatmap of dataset
def correlation_heatmap(df):_ , ax = plt.subplots(figsize =(14, 12))colormap = sns.diverging_palette(220, 10, as_cmap = True)_ = sns.heatmap(df.corr(), cmap = colormap,square=True, cbar_kws={'shrink':.9 }, ax=ax,annot=True, linewidths=0.1,vmax=1.0, linecolor='white',annot_kws={'fontsize':12 })plt.title('Pearson Correlation of Features', y=1.05, size=15)correlation_heatmap(data1)结果:
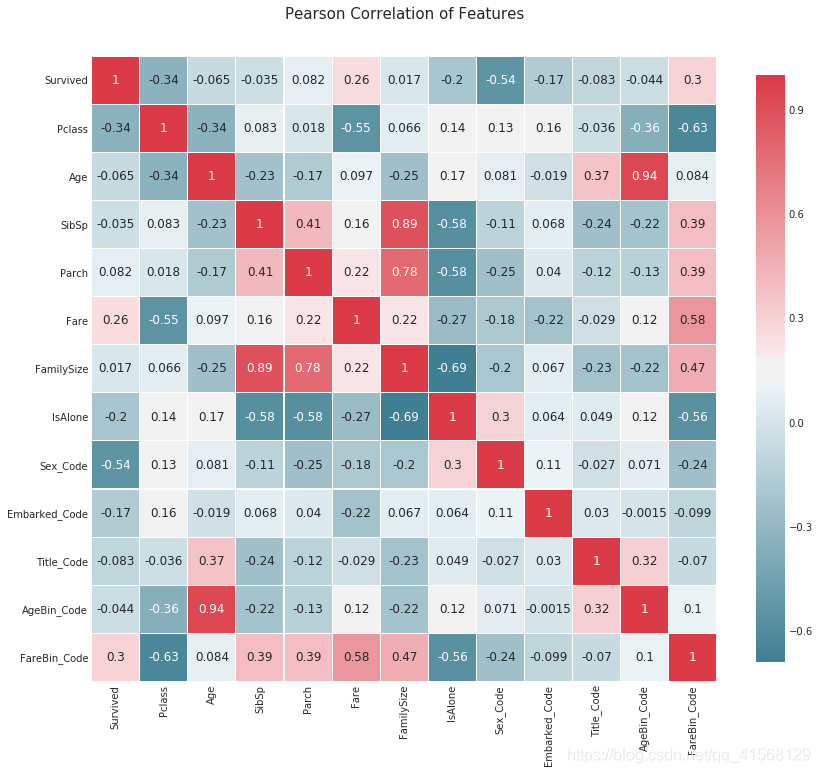
2.5 数据建模
与描述性统计和推断统计一样,数据建模既可以总结数据,也可以预测未来的结果。您的数据集和预期结果将决定可用的算法。重要的是要记住,算法只是工具,而不是魔法棒。你必须仍然是精通工艺的人,知道如何为工作选择合适的工具。一个类比就是让别人给你一把Philip螺丝刀,然后他们给你一把平头螺丝刀,或者最糟糕的是一把锤子。最坏的情况是,这使你不能完成这项任务。数据建模也是如此。错误的模型会导致糟糕的性能,而最坏的结果是错误的结论(这将被用作智能操作)。
数据科学是数学(即统计学、线性代数等)、计算机科学(即编程语言、计算机系统等)和商业管理(即通信、项目管理等)之间的多学科领域。大多数数据科学家来自三个领域中的一个,所以他们倾向于倾向于这一学科。
Machine Learning (ML) is teaching the machine how-to think and not what to think.
机器学习的目的是解决人类的问题。机器学习可分为:监督学习、无监督学习和强化学习。监督学习是模型有Label。无监督学习没有。强化学习是前两种模式的混合,没有立即给出正确答案,而是在随后的一系列事件之后,进行强化学习。我们正在进行有监督的机器学习,因为我们通过向算法提供一组特性及其相应的目标来训练它。然后,我们希望从同一个数据集中为它提供一个新的子集,并在预测准确性方面有类似的结果。
有许多机器学习算法,但是它们可以简化为四个类别:分类、回归、聚类或降维,这取决于你的目标变量和数据建模目标。今天将重点放在分类和回归上,我们可以归纳为连续目标变量需要回归算法,离散目标变量需要分类算法。另一个需要注意的是,逻辑回归虽然名字中有回归,但实际上是一种分类算法。由于我们的问题是预测乘客是否幸存,这是一个离散的目标变量。我们将使用sklearn库中的分类算法开始分析。我们将使用交叉验证和评分指标(在后面的章节中讨论)对算法的性能进行排名和比较。
Machine Learning Selection:
- Sklearn Estimator Overview
- Sklearn Estimator Detail
- Choosing Estimator Mind Map
- Choosing Estimator Cheat Sheet
Machine Learning Classification Algorithms:
- Ensemble Methods
- Generalized Linear Models (GLM)
- Naive Bayes
- Nearest Neighbors
- Support Vector Machines (SVM)
- Decision Trees
- Discriminant Analysis
2.5.1 怎样选择算法
当涉及到数据建模时,初学者的问题总是,“什么是最好的机器学习算法?”对于这一点,初学者必须学习机器学习的无免费午餐定理(No Free Lunch Theorem)。简言之,NFLT指出,对于所有数据集,没有一种超级算法在所有情况下都能发挥最佳效果。因此,最好的方法是尝试多个MLA,对它们进行优化,并根据您的特定场景对它们进行比较。有鉴于此,已经做了一些很好的研究来比较算法,例如Caruana和Niculescu Mizil 2006,观看MLA比较的视频讲座。Ogutu,2011用于基因组选择。Fernandez Delgado et al,2014对比17个家庭的179个分类器,Thoma 2016 sklearn对比,还有一个学派认为,更多的数据胜过更好的算法。
有了这些信息,初学者从哪里开始呢?我建议从Trees, Bagging, Random Forests, and Boosting开始。它们基本上是决策树的不同实现,这是最容易学习和理解的概念。在下一节中讨论的,它们也比SVC更容易调优。下面,我将简要介绍如何运行和比较几个MLA,但是这个kernel的其余部分将集中在通过决策树及其派生工具学习数据建模。
#Machine Learning Algorithm (MLA) Selection and Initialization
MLA = [#Ensemble Methodsensemble.AdaBoostClassifier(),ensemble.BaggingClassifier(),ensemble.ExtraTreesClassifier(),ensemble.GradientBoostingClassifier(),ensemble.RandomForestClassifier(),#Gaussian Processesgaussian_process.GaussianProcessClassifier(),#GLMlinear_model.LogisticRegressionCV(),linear_model.PassiveAggressiveClassifier(),linear_model.RidgeClassifierCV(),linear_model.SGDClassifier(),linear_model.Perceptron(),#Navies Bayesnaive_bayes.BernoulliNB(),naive_bayes.GaussianNB(),#Nearest Neighborneighbors.KNeighborsClassifier(),#SVMsvm.SVC(probability=True),svm.NuSVC(probability=True),svm.LinearSVC(),#Trees tree.DecisionTreeClassifier(),tree.ExtraTreeClassifier(),#Discriminant Analysisdiscriminant_analysis.LinearDiscriminantAnalysis(),discriminant_analysis.QuadraticDiscriminantAnalysis(),#xgboost: http://xgboost.readthedocs.io/en/latest/model.htmlXGBClassifier() ]#split dataset in cross-validation with this splitter class: http://scikit-learn.org/stable/modules/generated/sklearn.model_selection.ShuffleSplit.html#sklearn.model_selection.ShuffleSplit
#note: this is an alternative to train_test_split
cv_split = model_selection.ShuffleSplit(n_splits = 10, test_size = .3, train_size = .6, random_state = 0 ) # run model 10x with 60/30 split intentionally leaving out 10%#create table to compare MLA metrics
MLA_columns = ['MLA Name', 'MLA Parameters','MLA Train Accuracy Mean', 'MLA Test Accuracy Mean', 'MLA Test Accuracy 3*STD' ,'MLA Time']
MLA_compare = pd.DataFrame(columns = MLA_columns)#create table to compare MLA predictions
MLA_predict = data1[Target]#index through MLA and save performance to table
row_index = 0
for alg in MLA:#set name and parametersMLA_name = alg.__class__.__name__MLA_compare.loc[row_index, 'MLA Name'] = MLA_nameMLA_compare.loc[row_index, 'MLA Parameters'] = str(alg.get_params())#score model with cross validation: http://scikit-learn.org/stable/modules/generated/sklearn.model_selection.cross_validate.html#sklearn.model_selection.cross_validatecv_results = model_selection.cross_validate(alg, data1[data1_x_bin], data1[Target], cv = cv_split,return_train_score = True)MLA_compare.loc[row_index, 'MLA Time'] = cv_results['fit_time'].mean()MLA_compare.loc[row_index, 'MLA Train Accuracy Mean'] = cv_results['train_score'].mean()MLA_compare.loc[row_index, 'MLA Test Accuracy Mean'] = cv_results['test_score'].mean() #if this is a non-bias random sample, then +/-3 standard deviations (std) from the mean, should statistically capture 99.7% of the subsetsMLA_compare.loc[row_index, 'MLA Test Accuracy 3*STD'] = cv_results['test_score'].std()*3 #let's know the worst that can happen!#save MLA predictions - see section 6 for usagealg.fit(data1[data1_x_bin], data1[Target])MLA_predict[MLA_name] = alg.predict(data1[data1_x_bin])row_index+=1#print and sort table: https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.sort_values.html
MLA_compare.sort_values(by = ['MLA Test Accuracy Mean'], ascending = False, inplace = True)
MLA_compare
#MLA_predict结果:
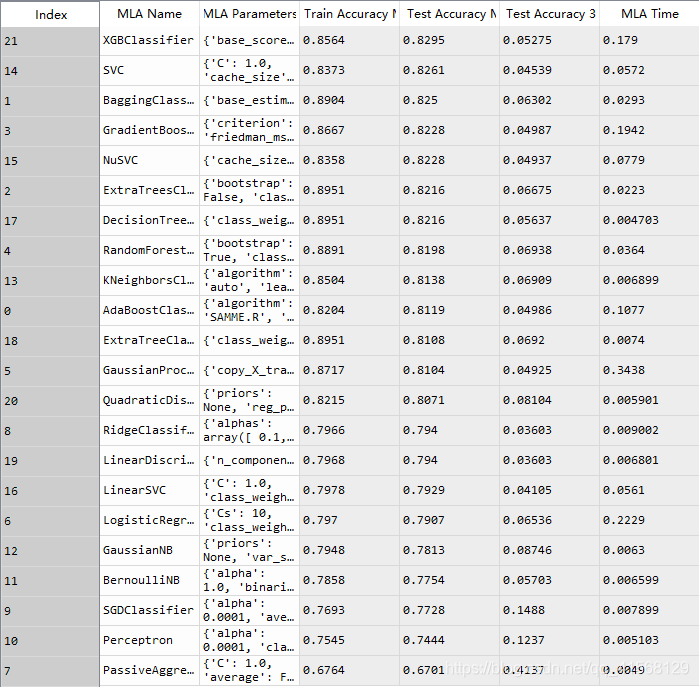
#barplot using https://seaborn.pydata.org/generated/seaborn.barplot.html
sns.barplot(x='MLA Test Accuracy Mean', y = 'MLA Name', data = MLA_compare, color = 'm')#prettify using pyplot: https://matplotlib.org/api/pyplot_api.html
plt.title('Machine Learning Algorithm Accuracy Score \n')
plt.xlabel('Accuracy Score (%)')
plt.ylabel('Algorithm')结果:
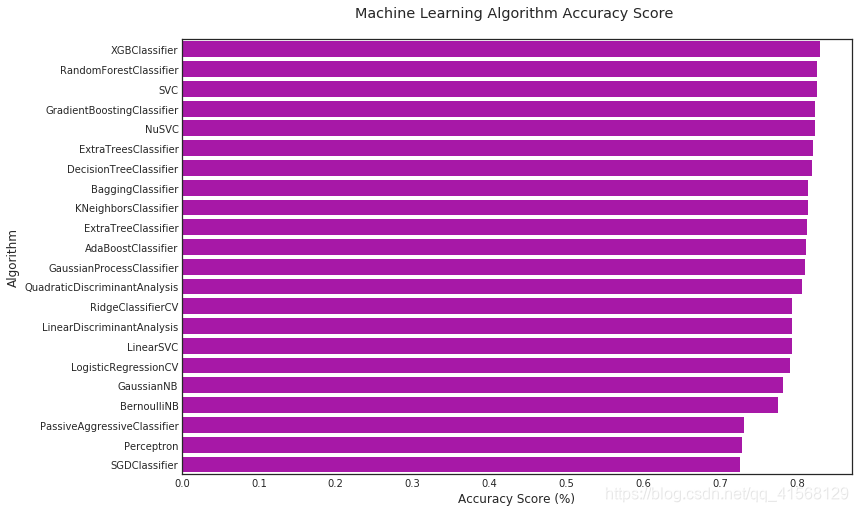
2.5.2 模型评估
到此,通过一些基本的数据清理、分析和机器学习算法(MLA),我们能够以大约82%的精度预测乘客的生存率。
制定最低正确率,我们知道1502/2224或67.5%的人死亡。因此,如果我们只是预测最常见的事件,即100%的人死亡,那么我们正确率应达到67.5%。所以把68%设为最差性能,低于此则不加考虑。
#group by or pivot table: https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.groupby.html
pivot_female = data1[data1.Sex=='female'].groupby(['Sex','Pclass', 'Embarked','FareBin'])['Survived'].mean()
print('Survival Decision Tree w/Female Node: \n',pivot_female)pivot_male = data1[data1.Sex=='male'].groupby(['Sex','Title'])['Survived'].mean()
print('\n\nSurvival Decision Tree w/Male Node: \n',pivot_male)结果:
Survival Decision Tree w/Female Node: Sex Pclass Embarked FareBin
female 1 C (14.454, 31.0] 0.666667(31.0, 512.329] 1.000000Q (31.0, 512.329] 1.000000S (14.454, 31.0] 1.000000(31.0, 512.329] 0.9555562 C (7.91, 14.454] 1.000000(14.454, 31.0] 1.000000(31.0, 512.329] 1.000000Q (7.91, 14.454] 1.000000S (7.91, 14.454] 0.875000(14.454, 31.0] 0.916667(31.0, 512.329] 1.0000003 C (-0.001, 7.91] 1.000000(7.91, 14.454] 0.428571(14.454, 31.0] 0.666667Q (-0.001, 7.91] 0.750000(7.91, 14.454] 0.500000(14.454, 31.0] 0.714286S (-0.001, 7.91] 0.533333(7.91, 14.454] 0.448276(14.454, 31.0] 0.357143(31.0, 512.329] 0.125000
Name: Survived, dtype: float64Survival Decision Tree w/Male Node: Sex Title
male Master 0.575000Misc 0.250000Mr 0.156673
Name: Survived, dtype: float64
#handmade data model using brain power (and Microsoft Excel Pivot Tables for quick calculations)
def mytree(df):#initialize table to store predictionsModel = pd.DataFrame(data = {'Predict':[]})male_title = ['Master'] #survived titlesfor index, row in df.iterrows():#Question 1: Were you on the Titanic; majority diedModel.loc[index, 'Predict'] = 0#Question 2: Are you female; majority survivedif (df.loc[index, 'Sex'] == 'female'):Model.loc[index, 'Predict'] = 1#Question 3A Female - Class and Question 4 Embarked gain minimum information#Question 5B Female - FareBin; set anything less than .5 in female node decision tree back to 0 if ((df.loc[index, 'Sex'] == 'female') & (df.loc[index, 'Pclass'] == 3) & (df.loc[index, 'Embarked'] == 'S') &(df.loc[index, 'Fare'] > 8)):Model.loc[index, 'Predict'] = 0#Question 3B Male: Title; set anything greater than .5 to 1 for majority survivedif ((df.loc[index, 'Sex'] == 'male') &(df.loc[index, 'Title'] in male_title)):Model.loc[index, 'Predict'] = 1return Model#model data
Tree_Predict = mytree(data1)
print('Decision Tree Model Accuracy/Precision Score: {:.2f}%\n'.format(metrics.accuracy_score(data1['Survived'], Tree_Predict)*100))#Accuracy Summary Report with http://scikit-learn.org/stable/modules/generated/sklearn.metrics.classification_report.html#sklearn.metrics.classification_report
#Where recall score = (true positives)/(true positive + false negative) w/1 being best:http://scikit-learn.org/stable/modules/generated/sklearn.metrics.recall_score.html#sklearn.metrics.recall_score
#And F1 score = weighted average of precision and recall w/1 being best: http://scikit-learn.org/stable/modules/generated/sklearn.metrics.f1_score.html#sklearn.metrics.f1_score
print(metrics.classification_report(data1['Survived'], Tree_Predict))结果:
Decision Tree Model Accuracy/Precision Score: 82.04%precision recall f1-score support0 0.82 0.91 0.86 5491 0.82 0.68 0.75 342avg / total 0.82 0.82 0.82 891
#Plot Accuracy Summary
#Credit: http://scikit-learn.org/stable/auto_examples/model_selection/plot_confusion_matrix.html
import itertools
def plot_confusion_matrix(cm, classes,normalize=False,title='Confusion matrix',cmap=plt.cm.Blues):"""This function prints and plots the confusion matrix.Normalization can be applied by setting `normalize=True`."""if normalize:cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]print("Normalized confusion matrix")else:print('Confusion matrix, without normalization')print(cm)plt.imshow(cm, interpolation='nearest', cmap=cmap)plt.title(title)plt.colorbar()tick_marks = np.arange(len(classes))plt.xticks(tick_marks, classes, rotation=45)plt.yticks(tick_marks, classes)fmt = '.2f' if normalize else 'd'thresh = cm.max() / 2.for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):plt.text(j, i, format(cm[i, j], fmt),horizontalalignment="center",color="white" if cm[i, j] > thresh else "black")plt.tight_layout()plt.ylabel('True label')plt.xlabel('Predicted label')# Compute confusion matrix
cnf_matrix = metrics.confusion_matrix(data1['Survived'], Tree_Predict)
np.set_printoptions(precision=2)class_names = ['Dead', 'Survived']
# Plot non-normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names,title='Confusion matrix, without normalization')# Plot normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names, normalize=True, title='Normalized confusion matrix')结果:
Confusion matrix, without normalization
[[497 52][108 234]]
Normalized confusion matrix
[[ 0.91 0.09][ 0.32 0.68]]

Cross-Validation (CV)检测模型性能
we used sklearn cross_validate function to train, test, and score our model performance.
2.5.3 使用超参数调优
class sklearn.tree.DecisionTreeClassifier(criterion=’gini’, splitter=’best’, max_depth=None, min_samples_split=2, min_samples_leaf=1, min_weight_fraction_leaf=0.0, max_features=None, random_state=None, max_leaf_nodes=None, min_impurity_decrease=0.0, min_impurity_split=None, class_weight=None, presort=False)
使用parametergrid、gridsearchcv和sklearn scoring来调整我们的模型;有关ROC_AUC的更多信息,用graphviz来可视化我们的树。
#base model
dtree = tree.DecisionTreeClassifier(random_state = 0)
base_results = model_selection.cross_validate(dtree, data1[data1_x_bin], data1[Target], cv = cv_split,return_train_score = True)
dtree.fit(data1[data1_x_bin], data1[Target])print('BEFORE DT Parameters: ', dtree.get_params())
print("BEFORE DT Training w/bin score mean: {:.2f}". format(base_results['train_score'].mean()*100))
print("BEFORE DT Test w/bin score mean: {:.2f}". format(base_results['test_score'].mean()*100))
print("BEFORE DT Test w/bin score 3*std: +/- {:.2f}". format(base_results['test_score'].std()*100*3))
#print("BEFORE DT Test w/bin set score min: {:.2f}". format(base_results['test_score'].min()*100))
print('-'*10)#tune hyper-parameters: http://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeClassifier.html#sklearn.tree.DecisionTreeClassifier
param_grid = {'criterion': ['gini', 'entropy'], #scoring methodology; two supported formulas for calculating information gain - default is gini#'splitter': ['best', 'random'], #splitting methodology; two supported strategies - default is best'max_depth': [2,4,6,8,10,None], #max depth tree can grow; default is none#'min_samples_split': [2,5,10,.03,.05], #minimum subset size BEFORE new split (fraction is % of total); default is 2#'min_samples_leaf': [1,5,10,.03,.05], #minimum subset size AFTER new split split (fraction is % of total); default is 1#'max_features': [None, 'auto'], #max features to consider when performing split; default none or all'random_state': [0] #seed or control random number generator: https://www.quora.com/What-is-seed-in-random-number-generation}#choose best model with grid_search: #http://scikit-learn.org/stable/modules/grid_search.html#grid-search
#http://scikit-learn.org/stable/auto_examples/model_selection/plot_grid_search_digits.html
tune_model = model_selection.GridSearchCV(tree.DecisionTreeClassifier(), param_grid=param_grid, scoring = 'roc_auc', cv = cv_split)
tune_model.fit(data1[data1_x_bin], data1[Target])#print(tune_model.cv_results_.keys())
#print(tune_model.cv_results_['params'])
print('AFTER DT Parameters: ', tune_model.best_params_)
#print(tune_model.cv_results_['mean_train_score'])
print("AFTER DT Training w/bin score mean: {:.2f}". format(tune_model.cv_results_['mean_train_score'][tune_model.best_index_]*100))
#print(tune_model.cv_results_['mean_test_score'])
print("AFTER DT Test w/bin score mean: {:.2f}". format(tune_model.cv_results_['mean_test_score'][tune_model.best_index_]*100))
print("AFTER DT Test w/bin score 3*std: +/- {:.2f}". format(tune_model.cv_results_['std_test_score'][tune_model.best_index_]*100*3))
print('-'*10)BEFORE DT Parameters: {'class_weight': None, 'criterion': 'gini', 'max_depth': None, 'max_features': None, 'max_leaf_nodes': None, 'min_impurity_decrease': 0.0, 'min_impurity_split': None, 'min_samples_leaf': 1, 'min_samples_split': 2, 'min_weight_fraction_leaf': 0.0, 'presort': False, 'random_state': 0, 'splitter': 'best'}
BEFORE DT Training w/bin score mean: 89.51
BEFORE DT Test w/bin score mean: 82.09
BEFORE DT Test w/bin score 3*std: +/- 5.57
----------
AFTER DT Parameters: {'criterion': 'gini', 'max_depth': 4, 'random_state': 0}
AFTER DT Training w/bin score mean: 89.35
AFTER DT Test w/bin score mean: 87.40
AFTER DT Test w/bin score 3*std: +/- 5.00
----------2.5.4 通过特征调整模型
如前所述,预测变量越多不意味模型越好,但正确的预测变量可以。所以数据建模的另一个步骤是特征选择。sklearn有几个选项,我们将使用递归特征消除(rfe)和交叉验证(cv)。
#base model
print('BEFORE DT RFE Training Shape Old: ', data1[data1_x_bin].shape)
print('BEFORE DT RFE Training Columns Old: ', data1[data1_x_bin].columns.values)print("BEFORE DT RFE Training w/bin score mean: {:.2f}". format(base_results['train_score'].mean()*100))
print("BEFORE DT RFE Test w/bin score mean: {:.2f}". format(base_results['test_score'].mean()*100))
print("BEFORE DT RFE Test w/bin score 3*std: +/- {:.2f}". format(base_results['test_score'].std()*100*3))
print('-'*10)#feature selection
dtree_rfe = feature_selection.RFECV(dtree, step = 1, scoring = 'accuracy', cv = cv_split)
dtree_rfe.fit(data1[data1_x_bin], data1[Target])#transform x&y to reduced features and fit new model
#alternative: can use pipeline to reduce fit and transform steps: http://scikit-learn.org/stable/modules/generated/sklearn.pipeline.Pipeline.html
X_rfe = data1[data1_x_bin].columns.values[dtree_rfe.get_support()]
rfe_results = model_selection.cross_validate(dtree, data1[X_rfe], data1[Target], cv = cv_split,return_train_score = True)#print(dtree_rfe.grid_scores_)
print('AFTER DT RFE Training Shape New: ', data1[X_rfe].shape)
print('AFTER DT RFE Training Columns New: ', X_rfe)print("AFTER DT RFE Training w/bin score mean: {:.2f}". format(rfe_results['train_score'].mean()*100))
print("AFTER DT RFE Test w/bin score mean: {:.2f}". format(rfe_results['test_score'].mean()*100))
print("AFTER DT RFE Test w/bin score 3*std: +/- {:.2f}". format(rfe_results['test_score'].std()*100*3))
print('-'*10)#tune rfe model
rfe_tune_model = model_selection.GridSearchCV(tree.DecisionTreeClassifier(), param_grid=param_grid, scoring = 'roc_auc', cv = cv_split)
rfe_tune_model.fit(data1[X_rfe], data1[Target])#print(rfe_tune_model.cv_results_.keys())
#print(rfe_tune_model.cv_results_['params'])
print('AFTER DT RFE Tuned Parameters: ', rfe_tune_model.best_params_)
#print(rfe_tune_model.cv_results_['mean_train_score'])
print("AFTER DT RFE Tuned Training w/bin score mean: {:.2f}". format(rfe_tune_model.cv_results_['mean_train_score'][tune_model.best_index_]*100))
#print(rfe_tune_model.cv_results_['mean_test_score'])
print("AFTER DT RFE Tuned Test w/bin score mean: {:.2f}". format(rfe_tune_model.cv_results_['mean_test_score'][tune_model.best_index_]*100))
print("AFTER DT RFE Tuned Test w/bin score 3*std: +/- {:.2f}". format(rfe_tune_model.cv_results_['std_test_score'][tune_model.best_index_]*100*3))
print('-'*10)结果:
BEFORE DT RFE Training Shape Old: (891, 7)
BEFORE DT RFE Training Columns Old: ['Sex_Code' 'Pclass' 'Embarked_Code' 'Title_Code' 'FamilySize''AgeBin_Code' 'FareBin_Code']
BEFORE DT RFE Training w/bin score mean: 89.51
BEFORE DT RFE Test w/bin score mean: 82.09
BEFORE DT RFE Test w/bin score 3*std: +/- 5.57
----------
AFTER DT RFE Training Shape New: (891, 6)
AFTER DT RFE Training Columns New: ['Sex_Code' 'Pclass' 'Title_Code' 'FamilySize' 'AgeBin_Code' 'FareBin_Code']
AFTER DT RFE Training w/bin score mean: 88.16
AFTER DT RFE Test w/bin score mean: 83.06
AFTER DT RFE Test w/bin score 3*std: +/- 6.22
----------
AFTER DT RFE Tuned Parameters: {'criterion': 'gini', 'max_depth': 4, 'random_state': 0}
AFTER DT RFE Tuned Training w/bin score mean: 89.39
AFTER DT RFE Tuned Test w/bin score mean: 87.34
AFTER DT RFE Tuned Test w/bin score 3*std: +/- 6.21
----------2.6 模型验证及实现
在您根据数据的一个子集对模型进行训练之后,是时候测试您的模型了。这有助于确保模型没有过拟合,或者其特定于所选子集,从而使其无法准确地适合同一数据集中的其他子集。在这一步中,我们将使用测试集确定模型是否过度拟合、归纳及欠拟合。
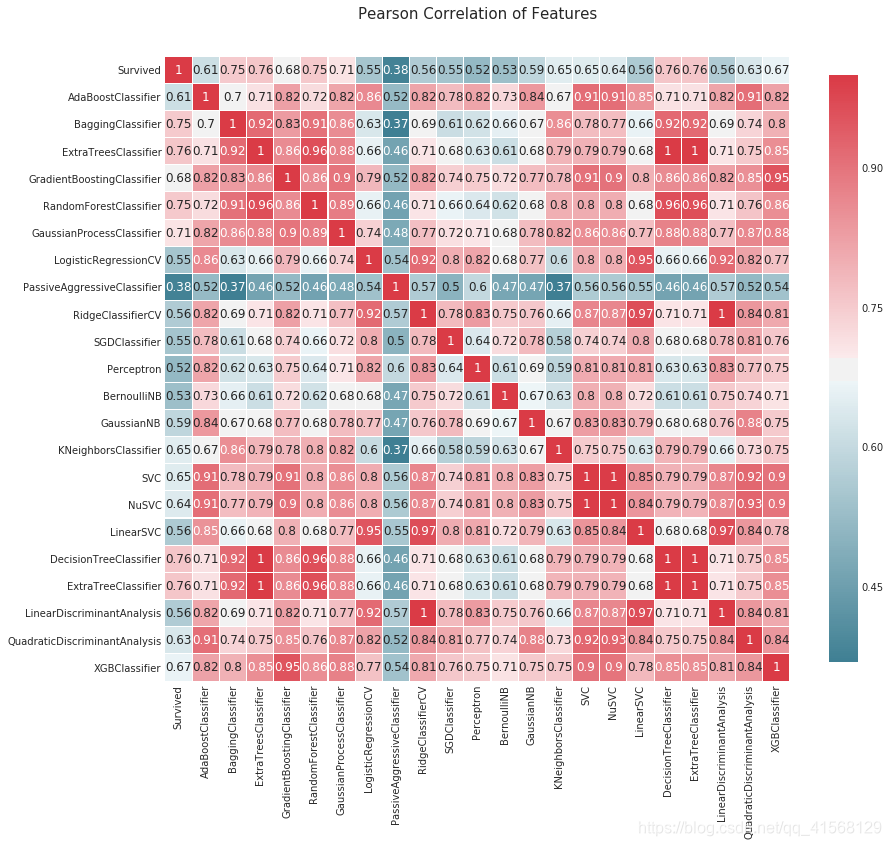
#compare algorithm predictions with each other, where 1 = exactly similar and 0 = exactly opposite
#there are some 1's, but enough blues and light reds to create a "super algorithm" by combining them
correlation_heatmap(MLA_predict)结果:
#why choose one model, when you can pick them all with voting classifier
#http://scikit-learn.org/stable/modules/generated/sklearn.ensemble.VotingClassifier.html
#removed models w/o attribute 'predict_proba' required for vote classifier and models with a 1.0 correlation to another model
vote_est = [#Ensemble Methods: http://scikit-learn.org/stable/modules/ensemble.html('ada', ensemble.AdaBoostClassifier()),('bc', ensemble.BaggingClassifier()),('etc',ensemble.ExtraTreesClassifier()),('gbc', ensemble.GradientBoostingClassifier()),('rfc', ensemble.RandomForestClassifier()),#Gaussian Processes: http://scikit-learn.org/stable/modules/gaussian_process.html#gaussian-process-classification-gpc('gpc', gaussian_process.GaussianProcessClassifier()),#GLM: http://scikit-learn.org/stable/modules/linear_model.html#logistic-regression('lr', linear_model.LogisticRegressionCV()),#Navies Bayes: http://scikit-learn.org/stable/modules/naive_bayes.html('bnb', naive_bayes.BernoulliNB()),('gnb', naive_bayes.GaussianNB()),#Nearest Neighbor: http://scikit-learn.org/stable/modules/neighbors.html('knn', neighbors.KNeighborsClassifier()),#SVM: http://scikit-learn.org/stable/modules/svm.html('svc', svm.SVC(probability=True)),#xgboost: http://xgboost.readthedocs.io/en/latest/model.html('xgb', XGBClassifier())]#Hard Vote or majority rules
vote_hard = ensemble.VotingClassifier(estimators = vote_est , voting = 'hard')
vote_hard_cv = model_selection.cross_validate(vote_hard, data1[data1_x_bin], data1[Target], cv = cv_split,return_train_score = True)
vote_hard.fit(data1[data1_x_bin], data1[Target])print("Hard Voting Training w/bin score mean: {:.2f}". format(vote_hard_cv['train_score'].mean()*100))
print("Hard Voting Test w/bin score mean: {:.2f}". format(vote_hard_cv['test_score'].mean()*100))
print("Hard Voting Test w/bin score 3*std: +/- {:.2f}". format(vote_hard_cv['test_score'].std()*100*3))
print('-'*10)#Soft Vote or weighted probabilities
vote_soft = ensemble.VotingClassifier(estimators = vote_est , voting = 'soft')
vote_soft_cv = model_selection.cross_validate(vote_soft, data1[data1_x_bin], data1[Target], cv = cv_split,return_train_score = True)
vote_soft.fit(data1[data1_x_bin], data1[Target])print("Soft Voting Training w/bin score mean: {:.2f}". format(vote_soft_cv['train_score'].mean()*100))
print("Soft Voting Test w/bin score mean: {:.2f}". format(vote_soft_cv['test_score'].mean()*100))
print("Soft Voting Test w/bin score 3*std: +/- {:.2f}". format(vote_soft_cv['test_score'].std()*100*3))
print('-'*10)结果:
Hard Voting Training w/bin score mean: 86.59
Hard Voting Test w/bin score mean: 82.39
Hard Voting Test w/bin score 3*std: +/- 4.95
----------
Soft Voting Training w/bin score mean: 87.15
Soft Voting Test w/bin score mean: 82.35
Soft Voting Test w/bin score 3*std: +/- 4.85
----------#WARNING: Running is very computational intensive and time expensive.
#Code is written for experimental/developmental purposes and not production ready!#Hyperparameter Tune with GridSearchCV: http://scikit-learn.org/stable/modules/generated/sklearn.model_selection.GridSearchCV.html
grid_n_estimator = [10, 50, 100, 300]
grid_ratio = [.1, .25, .5, .75, 1.0]
grid_learn = [.01, .03, .05, .1, .25]
grid_max_depth = [2, 4, 6, 8, 10, None]
grid_min_samples = [5, 10, .03, .05, .10]
grid_criterion = ['gini', 'entropy']
grid_bool = [True, False]
grid_seed = [0]grid_param = [[{#AdaBoostClassifier - http://scikit-learn.org/stable/modules/generated/sklearn.ensemble.AdaBoostClassifier.html'n_estimators': grid_n_estimator, #default=50'learning_rate': grid_learn, #default=1#'algorithm': ['SAMME', 'SAMME.R'], #default=’SAMME.R'random_state': grid_seed}],[{#BaggingClassifier - http://scikit-learn.org/stable/modules/generated/sklearn.ensemble.BaggingClassifier.html#sklearn.ensemble.BaggingClassifier'n_estimators': grid_n_estimator, #default=10'max_samples': grid_ratio, #default=1.0'random_state': grid_seed}],[{#ExtraTreesClassifier - http://scikit-learn.org/stable/modules/generated/sklearn.ensemble.ExtraTreesClassifier.html#sklearn.ensemble.ExtraTreesClassifier'n_estimators': grid_n_estimator, #default=10'criterion': grid_criterion, #default=”gini”'max_depth': grid_max_depth, #default=None'random_state': grid_seed}],[{#GradientBoostingClassifier - http://scikit-learn.org/stable/modules/generated/sklearn.ensemble.GradientBoostingClassifier.html#sklearn.ensemble.GradientBoostingClassifier#'loss': ['deviance', 'exponential'], #default=’deviance’'learning_rate': [.05], #default=0.1 -- 12/31/17 set to reduce runtime -- The best parameter for GradientBoostingClassifier is {'learning_rate': 0.05, 'max_depth': 2, 'n_estimators': 300, 'random_state': 0} with a runtime of 264.45 seconds.'n_estimators': [300], #default=100 -- 12/31/17 set to reduce runtime -- The best parameter for GradientBoostingClassifier is {'learning_rate': 0.05, 'max_depth': 2, 'n_estimators': 300, 'random_state': 0} with a runtime of 264.45 seconds.#'criterion': ['friedman_mse', 'mse', 'mae'], #default=”friedman_mse”'max_depth': grid_max_depth, #default=3 'random_state': grid_seed}],[{#RandomForestClassifier - http://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html#sklearn.ensemble.RandomForestClassifier'n_estimators': grid_n_estimator, #default=10'criterion': grid_criterion, #default=”gini”'max_depth': grid_max_depth, #default=None'oob_score': [True], #default=False -- 12/31/17 set to reduce runtime -- The best parameter for RandomForestClassifier is {'criterion': 'entropy', 'max_depth': 6, 'n_estimators': 100, 'oob_score': True, 'random_state': 0} with a runtime of 146.35 seconds.'random_state': grid_seed}],[{ #GaussianProcessClassifier'max_iter_predict': grid_n_estimator, #default: 100'random_state': grid_seed}],[{#LogisticRegressionCV - http://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegressionCV.html#sklearn.linear_model.LogisticRegressionCV'fit_intercept': grid_bool, #default: True#'penalty': ['l1','l2'],'solver': ['newton-cg', 'lbfgs', 'liblinear', 'sag', 'saga'], #default: lbfgs'random_state': grid_seed}],[{#BernoulliNB - http://scikit-learn.org/stable/modules/generated/sklearn.naive_bayes.BernoulliNB.html#sklearn.naive_bayes.BernoulliNB'alpha': grid_ratio, #default: 1.0}],#GaussianNB - [{}],[{#KNeighborsClassifier - http://scikit-learn.org/stable/modules/generated/sklearn.neighbors.KNeighborsClassifier.html#sklearn.neighbors.KNeighborsClassifier'n_neighbors': [1,2,3,4,5,6,7], #default: 5'weights': ['uniform', 'distance'], #default = ‘uniform’'algorithm': ['auto', 'ball_tree', 'kd_tree', 'brute']}],[{#SVC - http://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html#sklearn.svm.SVC#http://blog.hackerearth.com/simple-tutorial-svm-parameter-tuning-python-r#'kernel': ['linear', 'poly', 'rbf', 'sigmoid'],'C': [1,2,3,4,5], #default=1.0'gamma': grid_ratio, #edfault: auto'decision_function_shape': ['ovo', 'ovr'], #default:ovr'probability': [True],'random_state': grid_seed}],[{#XGBClassifier - http://xgboost.readthedocs.io/en/latest/parameter.html'learning_rate': grid_learn, #default: .3'max_depth': [1,2,4,6,8,10], #default 2'n_estimators': grid_n_estimator, 'seed': grid_seed }] ]start_total = time.perf_counter() #https://docs.python.org/3/library/time.html#time.perf_counter
for clf, param in zip (vote_est, grid_param): #https://docs.python.org/3/library/functions.html#zip#print(clf[1]) #vote_est is a list of tuples, index 0 is the name and index 1 is the algorithm#print(param)start = time.perf_counter() best_search = model_selection.GridSearchCV(estimator = clf[1], param_grid = param, cv = cv_split, scoring = 'roc_auc')best_search.fit(data1[data1_x_bin], data1[Target])run = time.perf_counter() - startbest_param = best_search.best_params_print('The best parameter for {} is {} with a runtime of {:.2f} seconds.'.format(clf[1].__class__.__name__, best_param, run))clf[1].set_params(**best_param) run_total = time.perf_counter() - start_total
print('Total optimization time was {:.2f} minutes.'.format(run_total/60))print('-'*10)结果:
The best parameter for AdaBoostClassifier is {'learning_rate': 0.1, 'n_estimators': 300, 'random_state': 0} with a runtime of 37.28 seconds.
The best parameter for BaggingClassifier is {'max_samples': 0.25, 'n_estimators': 300, 'random_state': 0} with a runtime of 33.04 seconds.
The best parameter for ExtraTreesClassifier is {'criterion': 'entropy', 'max_depth': 6, 'n_estimators': 100, 'random_state': 0} with a runtime of 68.93 seconds.
The best parameter for GradientBoostingClassifier is {'learning_rate': 0.05, 'max_depth': 2, 'n_estimators': 300, 'random_state': 0} with a runtime of 38.77 seconds.
The best parameter for RandomForestClassifier is {'criterion': 'entropy', 'max_depth': 6, 'n_estimators': 100, 'oob_score': True, 'random_state': 0} with a runtime of 84.14 seconds.
The best parameter for GaussianProcessClassifier is {'max_iter_predict': 10, 'random_state': 0} with a runtime of 6.19 seconds.
The best parameter for LogisticRegressionCV is {'fit_intercept': True, 'random_state': 0, 'solver': 'liblinear'} with a runtime of 9.40 seconds.
The best parameter for BernoulliNB is {'alpha': 0.1} with a runtime of 0.24 seconds.
The best parameter for GaussianNB is {} with a runtime of 0.05 seconds.
The best parameter for KNeighborsClassifier is {'algorithm': 'brute', 'n_neighbors': 7, 'weights': 'uniform'} with a runtime of 5.56 seconds.
The best parameter for SVC is {'C': 2, 'decision_function_shape': 'ovo', 'gamma': 0.1, 'probability': True, 'random_state': 0} with a runtime of 30.49 seconds.
The best parameter for XGBClassifier is {'learning_rate': 0.01, 'max_depth': 4, 'n_estimators': 300, 'seed': 0} with a runtime of 43.57 seconds.
Total optimization time was 5.96 minutes.
----------#Hard Vote or majority rules w/Tuned Hyperparameters
grid_hard = ensemble.VotingClassifier(estimators = vote_est , voting = 'hard')
grid_hard_cv = model_selection.cross_validate(grid_hard, data1[data1_x_bin], data1[Target], cv = cv_split,return_train_score = True)
grid_hard.fit(data1[data1_x_bin], data1[Target])print("Hard Voting w/Tuned Hyperparameters Training w/bin score mean: {:.2f}". format(grid_hard_cv['train_score'].mean()*100))
print("Hard Voting w/Tuned Hyperparameters Test w/bin score mean: {:.2f}". format(grid_hard_cv['test_score'].mean()*100))
print("Hard Voting w/Tuned Hyperparameters Test w/bin score 3*std: +/- {:.2f}". format(grid_hard_cv['test_score'].std()*100*3))
print('-'*10)#Soft Vote or weighted probabilities w/Tuned Hyperparameters
grid_soft = ensemble.VotingClassifier(estimators = vote_est , voting = 'soft')
grid_soft_cv = model_selection.cross_validate(grid_soft, data1[data1_x_bin], data1[Target], cv = cv_split,return_train_score = True)
grid_soft.fit(data1[data1_x_bin], data1[Target])print("Soft Voting w/Tuned Hyperparameters Training w/bin score mean: {:.2f}". format(grid_soft_cv['train_score'].mean()*100))
print("Soft Voting w/Tuned Hyperparameters Test w/bin score mean: {:.2f}". format(grid_soft_cv['test_score'].mean()*100))
print("Soft Voting w/Tuned Hyperparameters Test w/bin score 3*std: +/- {:.2f}". format(grid_soft_cv['test_score'].std()*100*3))
print('-'*10)结果:
Hard Voting w/Tuned Hyperparameters Training w/bin score mean: 85.22
Hard Voting w/Tuned Hyperparameters Test w/bin score mean: 82.31
Hard Voting w/Tuned Hyperparameters Test w/bin score 3*std: +/- 5.26
----------
Soft Voting w/Tuned Hyperparameters Training w/bin score mean: 84.76
Soft Voting w/Tuned Hyperparameters Test w/bin score mean: 82.28
Soft Voting w/Tuned Hyperparameters Test w/bin score 3*std: +/- 5.42
----------
#prepare data for modeling
print(data_val.info())
print("-"*10)
#data_val.sample(10)#decision tree w/full dataset modeling submission score: defaults= 0.76555, tuned= 0.77990
#submit_dt = tree.DecisionTreeClassifier()
#submit_dt = model_selection.GridSearchCV(tree.DecisionTreeClassifier(), param_grid=param_grid, scoring = 'roc_auc', cv = cv_split)
#submit_dt.fit(data1[data1_x_bin], data1[Target])
#print('Best Parameters: ', submit_dt.best_params_) #Best Parameters: {'criterion': 'gini', 'max_depth': 4, 'random_state': 0}
#data_val['Survived'] = submit_dt.predict(data_val[data1_x_bin])#bagging w/full dataset modeling submission score: defaults= 0.75119, tuned= 0.77990
#submit_bc = ensemble.BaggingClassifier()
#submit_bc = model_selection.GridSearchCV(ensemble.BaggingClassifier(), param_grid= {'n_estimators':grid_n_estimator, 'max_samples': grid_ratio, 'oob_score': grid_bool, 'random_state': grid_seed}, scoring = 'roc_auc', cv = cv_split)
#submit_bc.fit(data1[data1_x_bin], data1[Target])
#print('Best Parameters: ', submit_bc.best_params_) #Best Parameters: {'max_samples': 0.25, 'n_estimators': 500, 'oob_score': True, 'random_state': 0}
#data_val['Survived'] = submit_bc.predict(data_val[data1_x_bin])#extra tree w/full dataset modeling submission score: defaults= 0.76555, tuned= 0.77990
#submit_etc = ensemble.ExtraTreesClassifier()
#submit_etc = model_selection.GridSearchCV(ensemble.ExtraTreesClassifier(), param_grid={'n_estimators': grid_n_estimator, 'criterion': grid_criterion, 'max_depth': grid_max_depth, 'random_state': grid_seed}, scoring = 'roc_auc', cv = cv_split)
#submit_etc.fit(data1[data1_x_bin], data1[Target])
#print('Best Parameters: ', submit_etc.best_params_) #Best Parameters: {'criterion': 'entropy', 'max_depth': 6, 'n_estimators': 100, 'random_state': 0}
#data_val['Survived'] = submit_etc.predict(data_val[data1_x_bin])#random foreset w/full dataset modeling submission score: defaults= 0.71291, tuned= 0.73205
#submit_rfc = ensemble.RandomForestClassifier()
#submit_rfc = model_selection.GridSearchCV(ensemble.RandomForestClassifier(), param_grid={'n_estimators': grid_n_estimator, 'criterion': grid_criterion, 'max_depth': grid_max_depth, 'random_state': grid_seed}, scoring = 'roc_auc', cv = cv_split)
#submit_rfc.fit(data1[data1_x_bin], data1[Target])
#print('Best Parameters: ', submit_rfc.best_params_) #Best Parameters: {'criterion': 'entropy', 'max_depth': 6, 'n_estimators': 100, 'random_state': 0}
#data_val['Survived'] = submit_rfc.predict(data_val[data1_x_bin])#ada boosting w/full dataset modeling submission score: defaults= 0.74162, tuned= 0.75119
#submit_abc = ensemble.AdaBoostClassifier()
#submit_abc = model_selection.GridSearchCV(ensemble.AdaBoostClassifier(), param_grid={'n_estimators': grid_n_estimator, 'learning_rate': grid_ratio, 'algorithm': ['SAMME', 'SAMME.R'], 'random_state': grid_seed}, scoring = 'roc_auc', cv = cv_split)
#submit_abc.fit(data1[data1_x_bin], data1[Target])
#print('Best Parameters: ', submit_abc.best_params_) #Best Parameters: {'algorithm': 'SAMME.R', 'learning_rate': 0.1, 'n_estimators': 300, 'random_state': 0}
#data_val['Survived'] = submit_abc.predict(data_val[data1_x_bin])#gradient boosting w/full dataset modeling submission score: defaults= 0.75119, tuned= 0.77033
#submit_gbc = ensemble.GradientBoostingClassifier()
#submit_gbc = model_selection.GridSearchCV(ensemble.GradientBoostingClassifier(), param_grid={'learning_rate': grid_ratio, 'n_estimators': grid_n_estimator, 'max_depth': grid_max_depth, 'random_state':grid_seed}, scoring = 'roc_auc', cv = cv_split)
#submit_gbc.fit(data1[data1_x_bin], data1[Target])
#print('Best Parameters: ', submit_gbc.best_params_) #Best Parameters: {'learning_rate': 0.25, 'max_depth': 2, 'n_estimators': 50, 'random_state': 0}
#data_val['Survived'] = submit_gbc.predict(data_val[data1_x_bin])#extreme boosting w/full dataset modeling submission score: defaults= 0.73684, tuned= 0.77990
#submit_xgb = XGBClassifier()
#submit_xgb = model_selection.GridSearchCV(XGBClassifier(), param_grid= {'learning_rate': grid_learn, 'max_depth': [0,2,4,6,8,10], 'n_estimators': grid_n_estimator, 'seed': grid_seed}, scoring = 'roc_auc', cv = cv_split)
#submit_xgb.fit(data1[data1_x_bin], data1[Target])
#print('Best Parameters: ', submit_xgb.best_params_) #Best Parameters: {'learning_rate': 0.01, 'max_depth': 4, 'n_estimators': 300, 'seed': 0}
#data_val['Survived'] = submit_xgb.predict(data_val[data1_x_bin])#handmade decision tree - submission score = 0.77990
data_val['Survived'] = mytree(data_val).astype(int)#hard voting classifier w/full dataset modeling submission score: defaults= 0.75598, tuned = 0.77990
#data_val['Survived'] = vote_hard.predict(data_val[data1_x_bin])
data_val['Survived'] = grid_hard.predict(data_val[data1_x_bin])#soft voting classifier w/full dataset modeling submission score: defaults= 0.73684, tuned = 0.74162
#data_val['Survived'] = vote_soft.predict(data_val[data1_x_bin])
#data_val['Survived'] = grid_soft.predict(data_val[data1_x_bin])#submit file
submit = data_val[['PassengerId','Survived']]
submit.to_csv("../working/submit.csv", index=False)print('Validation Data Distribution: \n', data_val['Survived'].value_counts(normalize = True))
submit.sample(10)结果:
RangeIndex: 418 entries, 0 to 417
Data columns (total 21 columns):
PassengerId 418 non-null int64
Pclass 418 non-null int64
Name 418 non-null object
Sex 418 non-null object
Age 418 non-null float64
SibSp 418 non-null int64
Parch 418 non-null int64
Ticket 418 non-null object
Fare 418 non-null float64
Cabin 91 non-null object
Embarked 418 non-null object
FamilySize 418 non-null int64
IsAlone 418 non-null int64
Title 418 non-null object
FareBin 418 non-null category
AgeBin 418 non-null category
Sex_Code 418 non-null int64
Embarked_Code 418 non-null int64
Title_Code 418 non-null int64
AgeBin_Code 418 non-null int64
FareBin_Code 418 non-null int64
dtypes: category(2), float64(2), int64(11), object(6)
memory usage: 63.1+ KB
None
----------
Validation Data Distribution: 0 0.633971
1 0.366029
Name: Survived, dtype: float64 
2.7 模型优化及策略
模型优化需要重复这个过程,使它变得更好…更强大…比以前更快。作为一名数据科学家,您的策略应该是将开发人员操作和应用程序管道外包,这样您就有更多的时间来关注建议和设计。一旦你能够包装你的想法,这就成为你的“兑换货币”。
原文链接
这篇关于Titanic: Machine Learning from Disaster的文章就介绍到这儿,希望我们推荐的文章对编程师们有所帮助!









