本文主要是介绍CryoEM - 冷冻电镜聚类中心(2D Class)粒子图像的解析,希望对大家解决编程问题提供一定的参考价值,需要的开发者们随着小编来一起学习吧!
欢迎关注我的CSDN:https://spike.blog.csdn.net/
本文地址:https://blog.csdn.net/caroline_wendy/article/details/126244406
冷冻电镜粒子图像聚类:
-
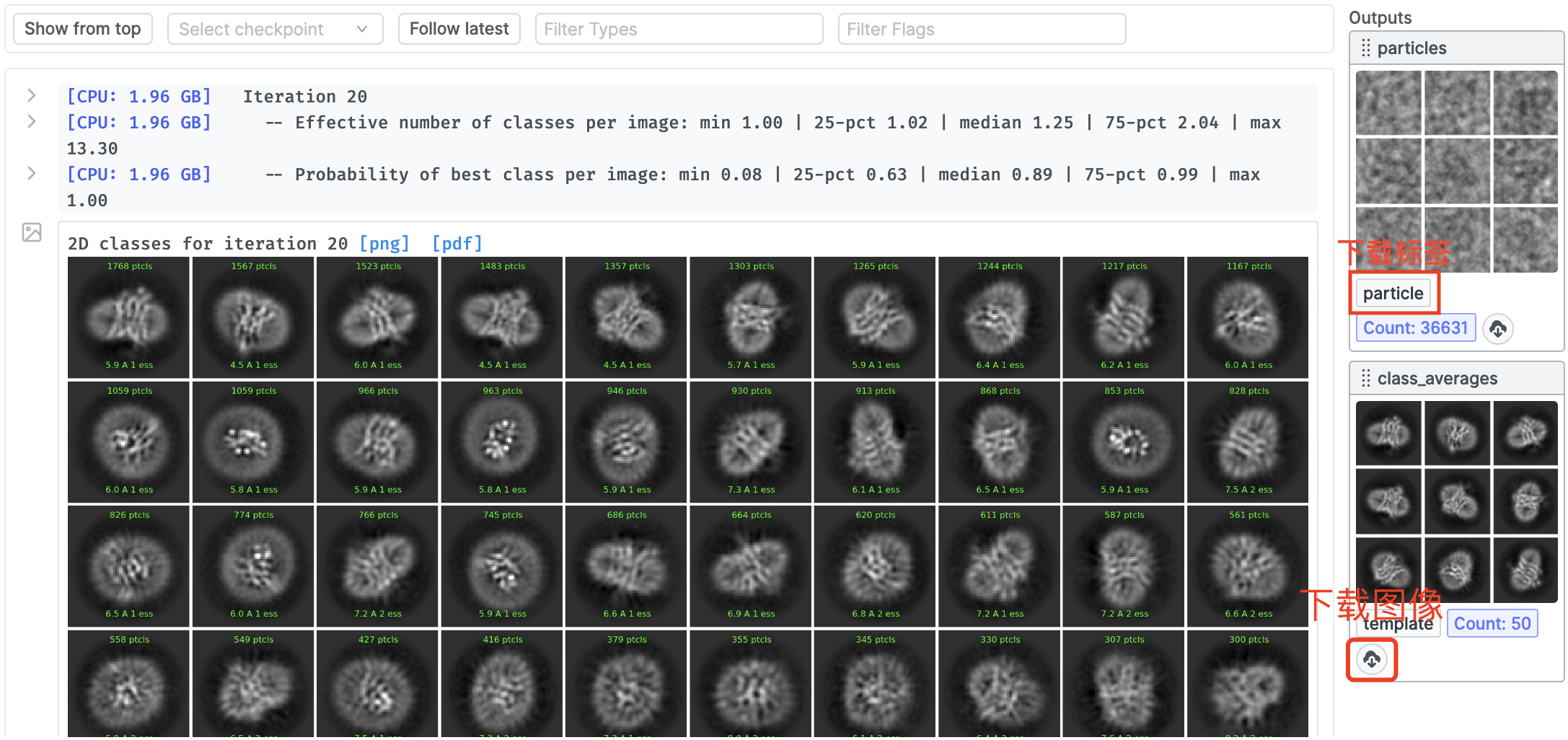
Import Particles:读取原始的冷冻电镜粒子图像
-
2D Class:2D聚类,获取聚类中心的清晰粒子图像

下载图像的 聚类中心(MRC文件) 和 标签信息(CS文件):
- MRC:
cryosparc_P1_J12_020_class_averages.mrc - CS:
cryosparc_P1_J12_020_particles.cs
加载MRC文件函数:
def mrc_loader(mrc_path):"""加载MRC文件"""mrc = mrcfile.open(mrc_path, permissive=True)data = mrc.dataarr = np.zeros(shape=data.shape, dtype=data.dtype)arr[:] = data[:]return arr
解析MRC文件:
def mrc_parser(mrc_path, out_dir, is_log=False):"""读取MRC文件,获取Particle图像的最小、最大值,归一化,存储于输出文件夹:param mrc_path: MRC文件:param out_dir: 输出文件夹:param is_log: 日志:return: None"""arr = mrc_loader(mrc_path)i_min, i_max = np.min(arr), np.max(arr)for i in tqdm(range(arr.shape[0]), desc="particles"):img = np.squeeze(arr[i, :, :])img = (img - i_min) / (i_max - i_min)img = np.clip(img * 255., 0, 255)img = img.astype(np.uint8)out_path = os.path.join(out_dir, "{}.png".format(i))cv2.imwrite(out_path, img)if is_log:print(f"[Info] arr: {arr.shape}, i_min: {i_min}, i_max: {i_max}")print(f"[Info] out_dir: {out_dir}")
样本:
| 0 | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
 |  |  |  |  |
安装PyEM包,用于处理CS文件:参考 Install pyem with Miniconda
conda activate cryosparc-master
conda install numpy scipy matplotlib seaborn numba pandas natsort
conda install -c conda-forge pyfftw healpy pathos # 安装速度较慢git clone https://github.com/asarnow/pyem.git
cd pyem
pip install --no-dependencies -e .
使用PyEM:参考 Export from cryoSPARC v2
运行脚本:
cd workspace/pyem
conda activate cryosparc-masterpython csparc2star.py cryosparc_P1_J12_020_particles.cs cryosparc_P1_J12_020_particles.star --relion2
数据,与待处理的particle图像,同名文件:
_rlnAnglePsi #2:面内旋转,PSI(ψ),Peptide torsion angles:肽扭转角_rlnClassNumber #15:类别,1~50- 样本从第20行开始,4个head行+15个label行
000001@J11/imported/017214887957000494328_000000855321499642015_stack_1293_cor2_DW_particles.mrc 83.571426 -0.450000 -5.250000 300.000000 10000.000000 1.082500 13015.895508 12950.861328 9.064259 2.700000 0.000000 0.100000 0.000000 29
000002@J11/imported/017214887957000494328_000000855321499642015_stack_1293_cor2_DW_particles.mrc 254.387726 -2.550000 1.050000 300.000000 10000.000000 1.082500 12611.307617 12546.273438 9.064259 2.700000 0.000000 0.100000 0.000000 15
测试,图像旋转效果:
- 原始路径:
datasets/cryoEM/kongfang/Data/extract - 替换路径:去掉索引017214887957000494328
Data/extract/000000855321499642015_stack_1293_cor2_DW_particles.mrc
28聚类中心角度差异较大:

测试Case:
000005@J11/imported/017214887957000494328_000000855321499642015_stack_1293_cor2_DW_particles.mrc 247.040802 -2.850000 0.750000 300.000000 10000.000000 1.082500 13292.802734 13227.768555 9.064259 2.700000 0.000000 0.100000 0.000000 28
000020@J11/imported/004046678624515756479_000077449707304157065_stack_2096_cor2_DW_particles.mrc 70.714287 -0.150000 0.750000 300.000000 10000.000000 1.082500 17615.394531 17506.113281 -3.673789 2.700000 0.000000 0.100000 0.000000 28
路径变换:
Data/extract/000000855321499642015_stack_1293_cor2_DW_particles.mrc
图像:
| raw | lowpass | rotated center |
|---|---|---|
 |  |  |
旋转聚类中心图像的源码:
def rotate(img, angle, out_wh=None):"""旋转粒子粒子图像,填充背景像素:param img: 待旋转的图像:param angle: 旋转角度,来源于_rlnAnglePsi,顺时针旋转:param out_wh: 输出尺寸:return: 旋转之后的图像"""h, w = img.shape[:2]bkg_val = int(np.argmax(np.bincount(img.flatten()))) # 背景像素,用于填充# 旋转图像img, _ = rotate_img_with_bound(img, 360-angle, border_value=bkg_val)# 截取图像中心img = center_crop(img, h, w)if out_wh:img = cv2.resize(img, out_wh)return img
这篇关于CryoEM - 冷冻电镜聚类中心(2D Class)粒子图像的解析的文章就介绍到这儿,希望我们推荐的文章对编程师们有所帮助!



