本文主要是介绍Histolab:病理切片的预处理工具|项目实战,希望对大家解决编程问题提供一定的参考价值,需要的开发者们随着小编来一起学习吧!
小罗碎碎念
今天这期推文主要介绍一个病理切片的预处理工具:Histolab。
通过这期推文你最终可以实现的效果如下:
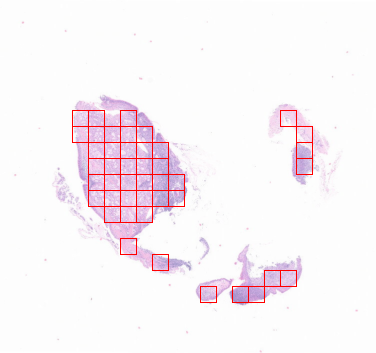
这只是一个简单的示意图,你看完本期推文以后可以自行调整。后期还会继续更新其他的模型,由易到难,现在看不上,小心以后跟不上哟,哈哈。
视频教程
复制网址,浏览器打开。
https://www.bilibili.com/video/BV1o6s4eoEwY/?vd_source=7c7c08bd51e5e86f3ee3a8766822236b
一、安装依赖项
随便打开一个虚拟环境,输入
pip install histolab
二、导入数据
2-1:确定路径
from histolab.slide import Slide
import os
BASE_PATH = os.getcwd()
print(BASE_PATH)
2-2:设置路径
CRC_path = '/data/data_mailab015/病理AI/人工智能病理/2024病理汇总-WDX/贵肿-svs/Raw-svs/1800518-1.svs' # location
path = r"./Histo" # 切割后图片存放的位置
CRC_slide = Slide(CRC_path,processed_path=path)
2-3:获取切片信息
print(f"Slide name:{CRC_slide.name}") # 幻灯片名称
print(f"Levels:{CRC_slide.levels}")
print(f"Dimensions at level 0:{CRC_slide.dimensions}")
print(f"Dimensions at level 1:{CRC_slide.level_dimensions(level=1)}")
print(f"Dimensions at level 2:{CRC_slide.level_dimensions(level=2)}")
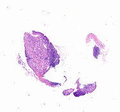
2-4:获取缩略图
CRC_slide.thumbnail
三、组织提取
3-1:随机提取
from histolab.tiler import RandomTilerrandom_tiles_extractor = RandomTiler(tile_size=(224, 224),n_tiles=30,level=0,seed=42,check_tissue=True, # defaulttissue_percent=80.0, # defaultprefix="random12/", # save tiles in the "random" subdirectory of slide's processed_pathsuffix=".png" # default
)
random_tiles_extractor.locate_tiles(slide=CRC_slide,scale_factor=24, # defaultalpha=128, # defaultoutline="red", # default
)
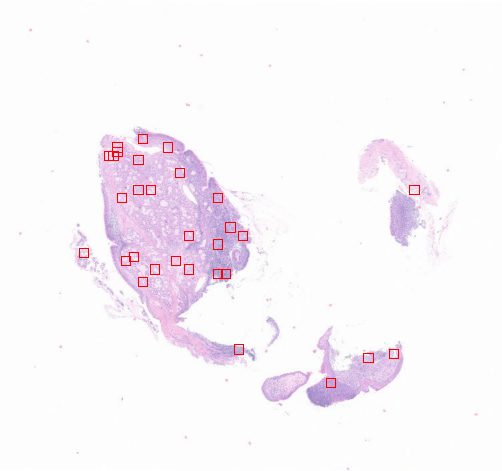
提取切片
random_tiles_extractor.extract(CRC_slide)
3-2:网格提取
from histolab.tiler import GridTilergrid_tiles_extractor = GridTiler(tile_size=(512, 512),level=0,check_tissue=False,pixel_overlap=0, # defaultprefix="grid/", # save tiles in the "grid" subdirectory of slide's processed_pathsuffix=".png" # default
)grid_tiles_extractor.locate_tiles(slide=CRC_slide,scale_factor=64,alpha=64,outline="#046C4C",
)
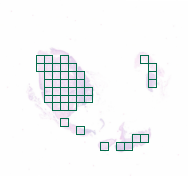
grid_tiles_extractor.extract(CRC_slide)
3-3:分数提取
from histolab.tiler import ScoreTiler
from histolab.scorer import NucleiScorer
scored_tiles_extractor = ScoreTiler(scorer = NucleiScorer(),tile_size=(512, 512),n_tiles=100,level=0,check_tissue=True,tissue_percent=80.0,pixel_overlap=0, # defaultprefix="scored/", # save tiles in the "scored" subdirectory of slide's processed_pathsuffix=".png" # default
)
grid_tiles_extractor.locate_tiles(slide=CRC_slide)

这篇关于Histolab:病理切片的预处理工具|项目实战的文章就介绍到这儿,希望我们推荐的文章对编程师们有所帮助!






