本文主要是介绍基因在各个细胞系表达情况,希望对大家解决编程问题提供一定的参考价值,需要的开发者们随着小编来一起学习吧!
从CCLE下载数据得到基因在每个细胞系中的
现在从DepMap: The Cancer Dependency Map Project at Broad Institute
需要先选择Custom Downloads

就可以下载数据进行处理了:
rm(list = ls())
library(tidyverse)
library(ggpubr)
rt <- data.table::fread("TP53 log2(TPM+1) Expression Public 23Q4.csv",data.table = F)##泛癌绘制##
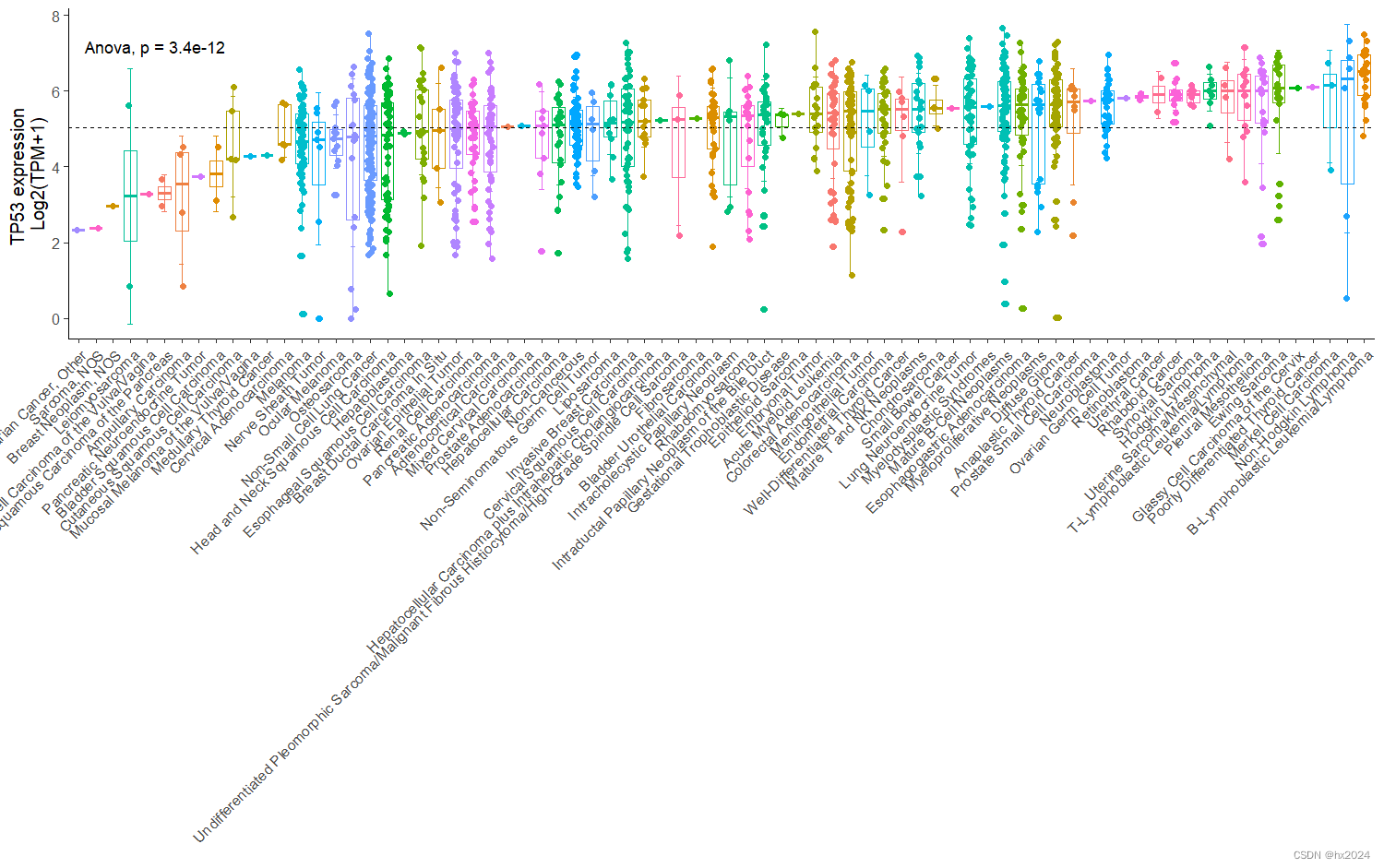
p1 <- ggplot(rt,aes(x=reorder(`Primary Disease`,`TP53 log2(TPM+1) Expression Public 23Q4`,FUN=median), #按中位数自动排序y=`TP53 log2(TPM+1) Expression Public 23Q4`,color=`Primary Disease`))+geom_boxplot()+geom_jitter(width = 0.15)+geom_hline(yintercept = mean(rt$`TP53 log2(TPM+1) Expression Public 23Q4`),lty=2)+theme_classic(base_size = 12)+rotate_x_text(45)+labs(x="",y="TP53 expression \nLog2(TPM+1)")+theme(legend.position = "none")+stat_summary(fun.data = "mean_sd",geom = "errorbar",width=0.3,position = position_dodge(0.9))+stat_compare_means(method = "anova",label.x = 3,label.y = 7)
p1
提取单个肿瘤:
##单个肿瘤##
dat1 <- rt[rt$`Primary Disease`=="Esophagogastric Adenocarcinoma",]
colnames(dat1)[2] <- "Expression Public"
p2 <- ggplot(dat1,aes(x=reorder(`Cell Line Name`,`Expression Public`,FUN=median), #按中位数自动排序y=`Expression Public`))+geom_segment(aes(y=mean(`Expression Public`),xend=`Cell Line Name`,yend=`Expression Public`))+geom_point(aes(size=`Expression Public`,color=`Expression Public`))+geom_hline(yintercept = mean(dat1$`Expression Public`),lty=2)+theme_bw(base_size = 12)+labs(x="",y="ALKBH5 expression",color="ALKBH5 expression",size="ALKBH5 expression")+scale_color_viridis_c(alpha = 1,begin = 0.6,end=0.9,direction = -1)+coord_flip()
p2
这篇关于基因在各个细胞系表达情况的文章就介绍到这儿,希望我们推荐的文章对编程师们有所帮助!






