本文主要是介绍RNA-seq 比对软件STAR——(2)使用,希望对大家解决编程问题提供一定的参考价值,需要的开发者们随着小编来一起学习吧!
RNA-seq 比对软件STAR——(2)使用
一、参数说明
详见——>manual
(1) readFilesIn
要映射序列文件的名称(带路径),注如果文件是压缩的文件使用readFilesCommand参数进行解压缩。如果是(*.gz)使用 --readFilesCommand zcat或 --readFilesCommand gunzip -c,对于bzip2压缩文件,使用–readFilesCommand bunzip2 -c
(2) outFileNamePrefix
输出文件的前缀(包含路径)
(3) outFilterMultimapNmax
一个read允许最多对齐数,超过认为read没有映射
max number of multiple alignments allowed for a read: if exceeded, the read is considered unmapped
(4) outSAMtype BAM SortedByCoordinate
生成的BAM文件排序
output sorted by coordinate Aligned.sortedByCoord.out.bam file, similar to samtools sort command. If this option causes problems, it is recommended to reduce
–outBAMsortingThreadN from the default 6 to lower values (as low as 1).
(5) outSAMattributes
- NH:number of loci the reads maps to: =1 for unique mappers, >1 for multimappers. Standard SAM tag.
- HI:multiple alignment index, starts with –outSAMattrIHstart (=1 by default). Standard SAM tag
- NM:edit distance to the reference (number of mismatched + inserted +deleted bases) for each mate. Standard SAM tag.
- MD:string encoding mismatched and deleted reference bases (see standard SAM specifications). Standard SAM tag.
- XS:alignment strand according to –outSAMstrandField.
- AS:multiple alignment index, starts with –outSAMattrIHstart (=1 by default). Standard SAM tag
二、index
STAR --runMode genomeGenerate --runThreadN 20 \
--genomeDir /share2/pub/yangjy/yangjy/database/STAR_index69 \
--outTmpDir /share2/pub/yangjy/yangjy/database/tmp \
--genomeFastaFiles /share/pub/wangxy/software/genome/ucsc/hg38/hg38.fa \
--sjdbGTFfile /share/pub/wangxy/Annotation/hg38/gencode.v34.annotation.gtf \
--sjdbOverhang 69
error 1
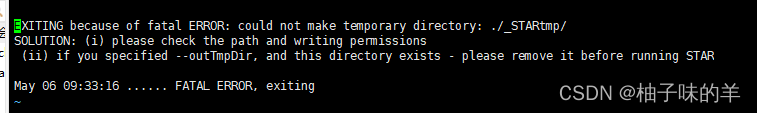
新版的STAR 需要写tmp路径,即增加参数 --outTmpDir ,而且这个路径必须不存在的!!,上面的STAR_index69必须是提前创建好的!!
error 2
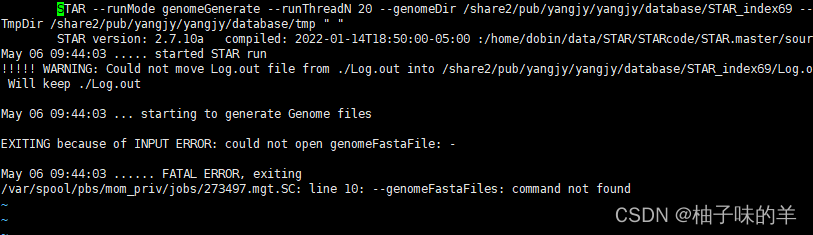
如果想要像我上面这种方式写脚本,一定要注意在每个反斜杠后面不能有空格或者其他字符!否则它认不得!其实可以直接写一行,但是为了方便看参数,我习惯这样写了,你们根据自己的习惯!
result
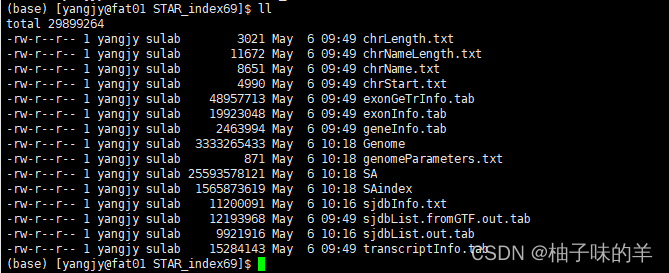
三、mapping
for file in 'SRR11296675' 'SRR11296676' 'SRR11296677' 'SRR11296678' 'SRR11296679' 'SRR11296680' 'SRR11296681' 'SRR11296682'
do
echo $file
STAR \
--runThreadN 40 \
--genomeDir /share2/pub/yangjy/yangjy/database/STAR_index69 \
--readFilesIn /share2/pub/yangjy/yangjy/rna-seq-data/GSE146887/fastq_data/$file.fastq \
--outFileNamePrefix /share2/pub/yangjy/yangjy/rna-seq-data/GSE146887/bbam/$file \
--outFilterMultimapNmax 500 \
--outSAMtype BAM SortedByCoordinate \
--outSAMattributes NH HI NM MD XS AS
done
result
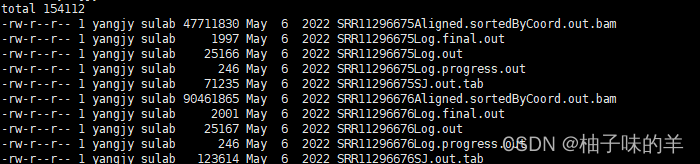
用过老版本的,新版本真的快很多很多~
这篇关于RNA-seq 比对软件STAR——(2)使用的文章就介绍到这儿,希望我们推荐的文章对编程师们有所帮助!





