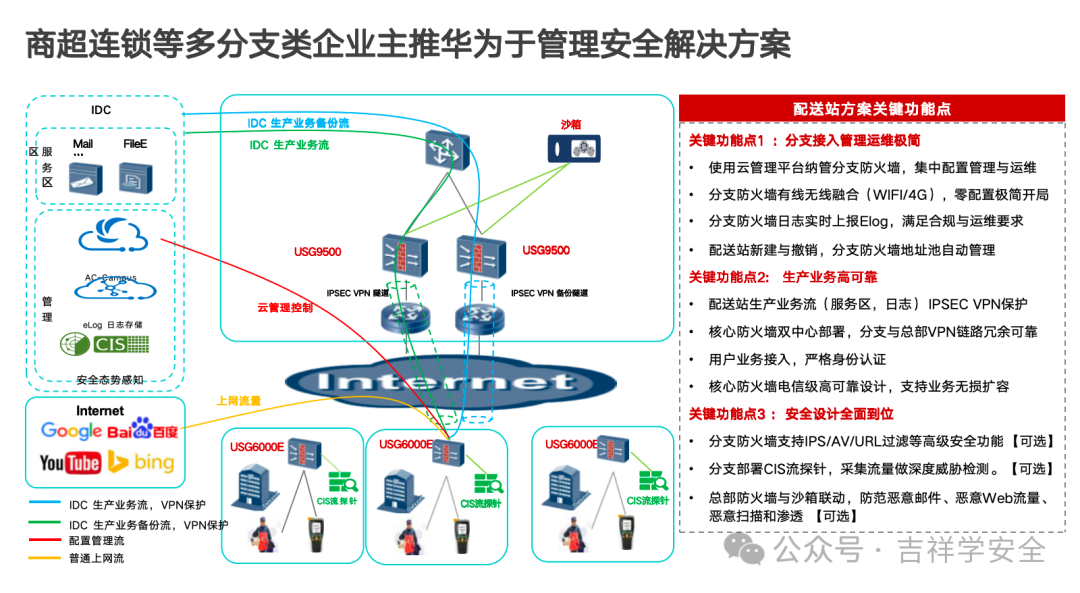
本文主要是介绍CellMarker | 人骨骼肌组织细胞Marker大全!~(强烈建议火速收藏!),希望对大家解决编程问题提供一定的参考价值,需要的开发者们随着小编来一起学习吧!
1写在前面
分享一下最近看到的2篇paper关于骨骼肌组织的细胞Marker,绝对的Atlas级好东西。👍
希望做单细胞的小伙伴觉得有用哦。😏
2常用marker(一)
general_mrkrs <- c(
'MYH7', 'TNNT1', 'TNNT3', 'MYH1', 'MYH2', "CKM", "MB", # Myofibers
'PAX7', 'DLK1', # MuSCs
'PDGFRA', 'DCN', 'ANGPTL7', 'OSR2', 'NGFR', 'SLC22A3','ITGA6', # Fibroblasts
'FMOD', 'TNMD' , 'MKX', # Tenocytes
'MPZ', 'MBP', # Schwann cells
'CDH2', 'L1CAM', # SCG
'MSLN', 'ITLN1', # mesothelium
"ADIPOQ", "PLIN1", # adipocytes
'PTPRC', 'CD3D', 'IL7R', # T cells
'NKG7', 'PRF1', #NK cells
'CD79A', "TCL1A", # B cells
'MZB1', 'JCHAIN', # B plasma
"CD14", "FCGR3A",'S100A8', 'S100A12', # Mono
"CD163", "C1QA", # Macrop
"XCR1", "CLEC9A", # cDC1 "CADM1",
"CD1C", "CLEC10A", "CCR7", # cDC2
'LILRA4', 'IL3RA', "IRF7", # pDC
'FCGR3B', 'CSF3R', 'SORL1', # Neutrophils
'EPX', 'PRG2', # Eosinophils 'CLC'
'TPSB2', 'MS4A2', # Mast cells
'PECAM1', 'HEY1','CLU', # art EC
'CA4', 'LPL', # capEC
'ACKR1', 'SELE', # venEC
'LYVE1', 'TFF3', # lymphEC
'RGS5','ABCC9', # pericytes
'MYH11', 'ACTA2', # SMC
'HBA1', #RBC
)
出自下面paper:👇
Human skeletal muscle aging atlas. Veronika R. Kedlian, Yaning Wang, Tianliang Liu, Xiaoping Chen, Liam Bolt, Catherine Tudor, Zhuojian Shen, Eirini S. Fasouli, Elena Prigmore, Vitalii Kleshchevnikov, Jan Patrick Pett, Tong Li, John E G Lawrence, Shani Perera, Martin Prete, Ni Huang, Qin Guo, Xinrui Zeng, Lu Yang, Krzysztof Polański, Nana-Jane Chipampe, Monika Dabrowska, Xiaobo Li, Omer Ali Bayraktar, Minal Patel, Natsuhiko Kumasaka, Krishnaa T. Mahbubani, Andy Peng Xiang, Kerstin B. Meyer, Kourosh Saeb-Parsy, Sarah A Teichmann & Hongbo Zhang 2024 Apr.
3常用marker(二)
Mural Cell Markers
#SMOOTH MUSCLE CELLS
FeaturePlot(df.harmony, features = "MYH11", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "ACTA2", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "TAGLN", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
#PERICYTES
FeaturePlot(df.harmony, features = "RGS5", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "CSPG4", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "PDGFRB", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
Glial Cells Markers
FeaturePlot(df.harmony, features = "PROX1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "MPZ", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "NCAM1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "CDH19", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "SOX10", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "PLP1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
Adipocites Markers
FeaturePlot(df.harmony, features = "PLIN1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "ADIPOQ", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "MMRN1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "CCL21", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
Tenocytes Markers
FeaturePlot(df.harmony, features = "FMOD", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "TNMD", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "COL22A1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "SCX", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "DLG2", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "FBN1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
Endothelial Markers
#FeaturePlot(df.harmony, features = "PCDHA6", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
#ARTERIAL
FeaturePlot(df.harmony, features = "FBLN5", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "DLL4", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "SEMA3G", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
#CAPILLARIES
FeaturePlot(df.harmony, features = "RGCC", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
#VENOUS
FeaturePlot(df.harmony, features = "EPHB4", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
Myonuclei Markers
FeaturePlot(df.harmony, features = "TTN", min.cutoff = "q9", order = T, cols = c("lightblue", "navy"), raster = FALSE)
#IMMATURE MYOCYTE
FeaturePlot(df.harmony, features = "MYMX", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "MYOG", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
#REG MYONUCLEI
FeaturePlot(df.harmony, features = "FLNC", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "MYH3", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "MYH8", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "XIRP1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
NMJ Myonuclei Markers (Neuromuscular junction)
#NMJ
FeaturePlot(df.harmony, features = "CHRNE", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "CHRNA1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "PRKAR1A", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "COL25A1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "UTRN", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "COLQ", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "ABLIM2", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "VAV3", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "UFSP1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
MTJ Myonuclei Markers (Myotendinous junction)
#MTJ
FeaturePlot(df.harmony, features = "COL22A1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "PIEZO2", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "COL24A1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "COL6A1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "FSTL1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "COL6A3", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "TIGD4", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(mini_df.harmony.harmony, features = "EYS", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
出自下面paper:👇
Lai, Y., Ramírez-Pardo, I., Isern, J. et al. Multimodal cell atlas of the ageing human skeletal muscle. Nature (2024).
点个在看吧各位~ ✐.ɴɪᴄᴇ ᴅᴀʏ 〰
📍 🤩 LASSO | 不来看看怎么美化你的LASSO结果吗!?
📍 🤣 chatPDF | 别再自己读文献了!让chatGPT来帮你读吧!~
📍 🤩 WGCNA | 值得你深入学习的生信分析方法!~
📍 🤩 ComplexHeatmap | 颜狗写的高颜值热图代码!
📍 🤥 ComplexHeatmap | 你的热图注释还挤在一起看不清吗!?
📍 🤨 Google | 谷歌翻译崩了我们怎么办!?(附完美解决方案)
📍 🤩 scRNA-seq | 吐血整理的单细胞入门教程
📍 🤣 NetworkD3 | 让我们一起画个动态的桑基图吧~
📍 🤩 RColorBrewer | 再多的配色也能轻松搞定!~
📍 🧐 rms | 批量完成你的线性回归
📍 🤩 CMplot | 完美复刻Nature上的曼哈顿图
📍 🤠 Network | 高颜值动态网络可视化工具
📍 🤗 boxjitter | 完美复刻Nature上的高颜值统计图
📍 🤫 linkET | 完美解决ggcor安装失败方案(附教程)
📍 ......




本文由 mdnice 多平台发布
这篇关于CellMarker | 人骨骼肌组织细胞Marker大全!~(强烈建议火速收藏!)的文章就介绍到这儿,希望我们推荐的文章对编程师们有所帮助!