本文主要是介绍单细胞UMAP降维图的修饰(一些偏僻问题),希望对大家解决编程问题提供一定的参考价值,需要的开发者们随着小编来一起学习吧!
本来呢,单细胞UMAP或者TSNE降维图正常展示就好了,最多修改颜色坐标就可以,但是奈何总是有文章挑战别人的眼睛,非要秀一波非常规操作,那我们也只能把他们打下神坛。
事情的起因是看了这篇文章中的UMAP图,有啥特点呢?每个点都是有边框的,这不就是ggplot中选择shape=21的形状吗?很显然,Dimplot是实现不了的。UMAP降维图本质也是散点图,只需要将作图数据导出,ggplot2就可以实现任何你想要的修饰了。

首先我们设置下颜色,并将作图的数据导出,导出的数据包含UMAP两个坐标的数据。
allcolour=c("#20B2AA","#FFA500","#9370DB","#98FB98","#1E90FF","#7CFC00","#FFFF00","#808000","#FF00FF","#FA8072","#7B68EE","#9400D3","#800080","#A0522D","#D2B48C","#D2691E","#87CEEB","#40E0D0","#5F9EA0","#FF1493","#0000CD","#008B8B","#FFE4B5","#8A2BE2","#228B22","#E9967A","#4682B4","#32CD32","#F0E68C","#FFFFE0","#EE82EE","#FF6347","#6A5ACD","#9932CC","#8B008B","#8B4513","#DEB887")
umap = NCsce@reductions$umap@cell.embeddings %>% as.data.frame() %>%cbind(cell_type = NCsce@meta.data$celltype)
然后就可以绘图了,很简单:
ggplot(umap, aes(x= UMAP_1 , y = UMAP_2 ,fill = cell_type))+geom_point(size = 3.5,colour="grey35",shape=21) +scale_fill_manual(values = allcolour)+ theme_classic()

可以像有些文章中一样,干脆不显示坐标轴,就是单纯的图。
ggplot(umap, aes(x= UMAP_1 , y = UMAP_2 ,fill = cell_type))+geom_point(size = 3,colour="grey35",shape=21) +scale_fill_manual(values = allcolour)+ theme_bw()+theme(panel.grid =element_blank(),panel.border = element_blank(),axis.text.y = element_blank(),axis.ticks.y = element_blank(),axis.ticks.x = element_blank(),axis.text.x = element_blank())+labs(x='', y='')
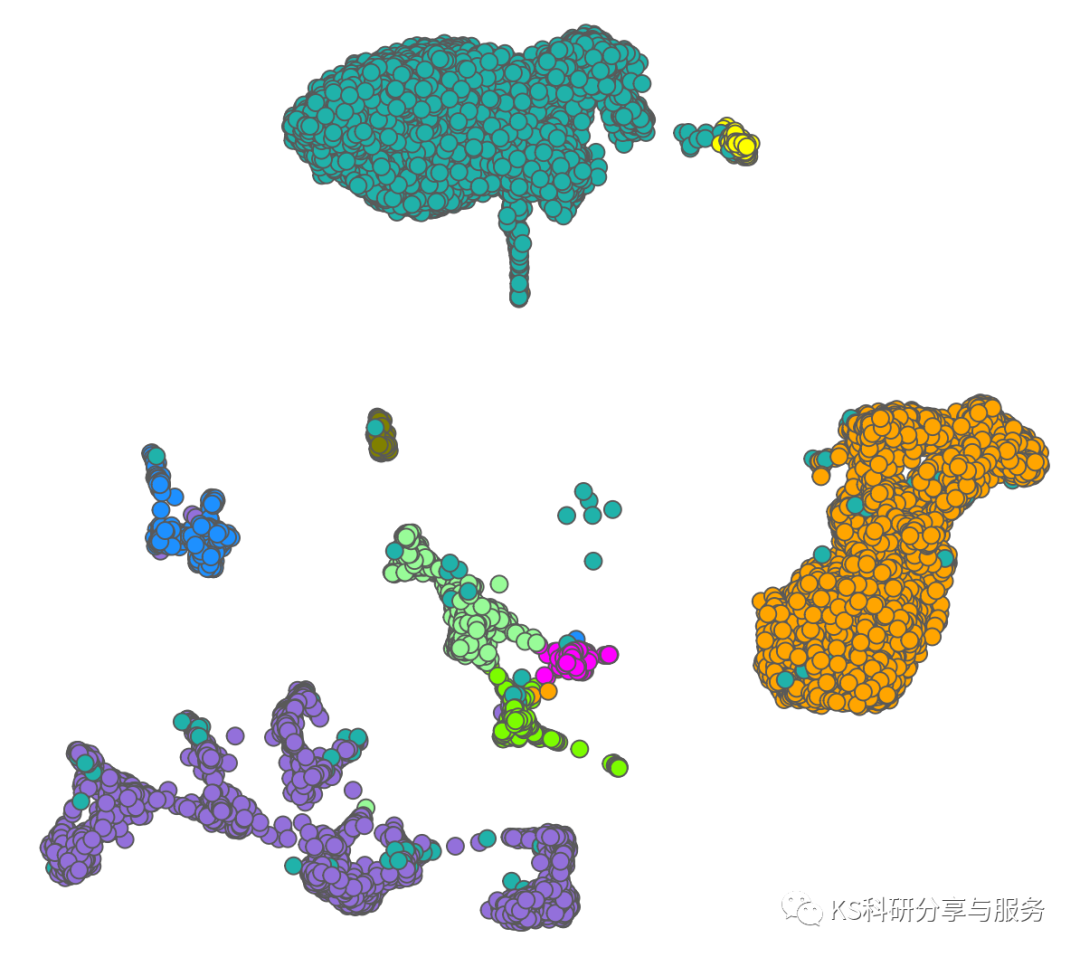
好了,这里也就是仅仅说明有这么一种情况存在,想要继续修饰可以发挥你们的聪明才智,争取有一个惊艳众人的可视化结果。
这篇关于单细胞UMAP降维图的修饰(一些偏僻问题)的文章就介绍到这儿,希望我们推荐的文章对编程师们有所帮助!








