本文主要是介绍PSP - 蛋白质与蛋白质的扩散对接 DiffDock-PP 算法,希望对大家解决编程问题提供一定的参考价值,需要的开发者们随着小编来一起学习吧!
欢迎关注我的CSDN:https://spike.blog.csdn.net/
本文地址:https://spike.blog.csdn.net/article/details/135115528
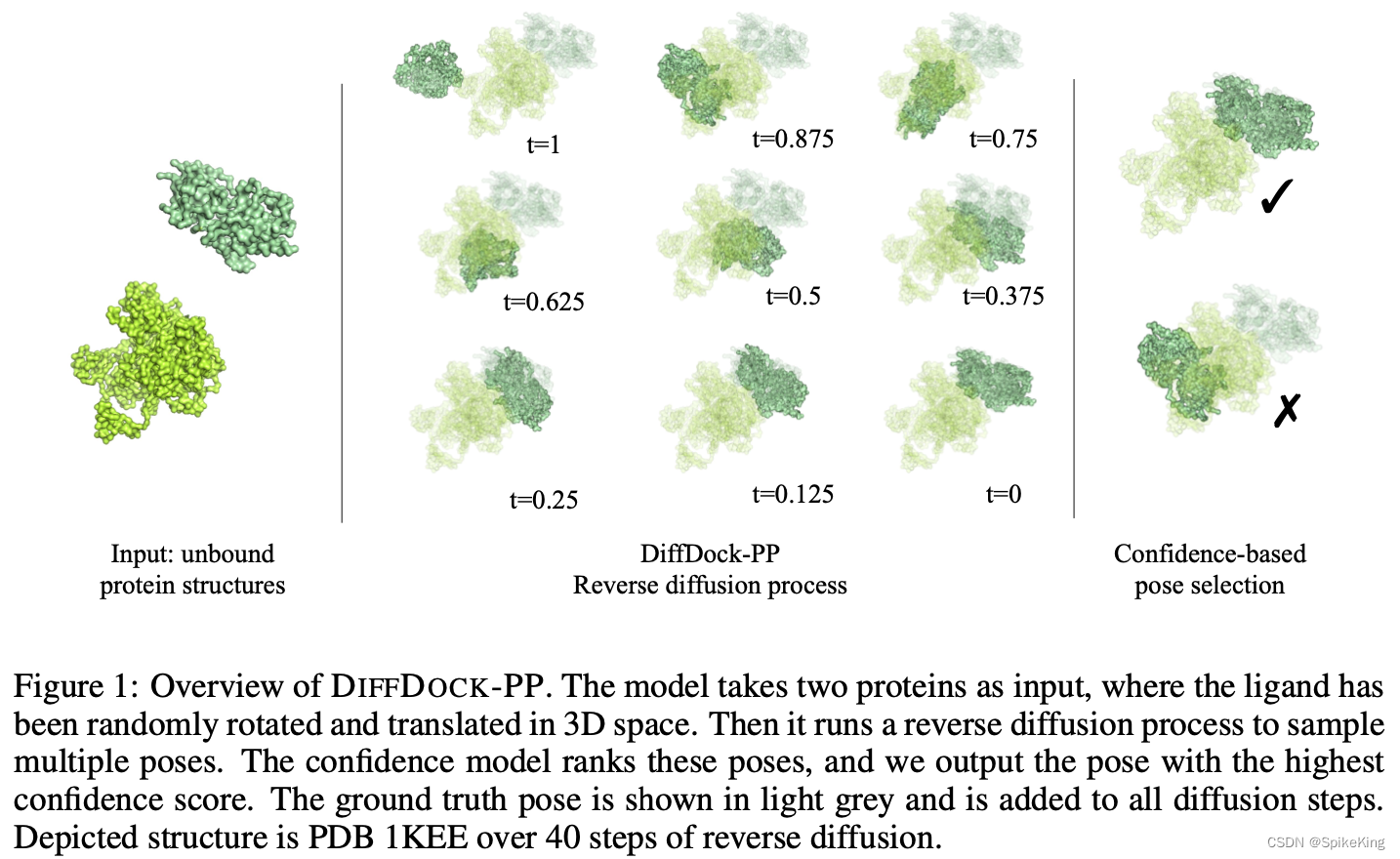
DiffDock-PP is a new approach to rigid-body protein-protein docking that is based on a diffusion generative model that learns to translate and rotate unbound protein structures into their bound conformations, and a confidence model that learns to rank different poses generated by the score model and select the best one.
DiffDock-PP 是一种新的刚体蛋白质-蛋白质对接方法,基于扩散生成模型,该模型学习将未结合的蛋白质结构翻译和旋转为其结合构象,基于置信模型,该模型学习对评分模型生成的不同姿势,进行排序并选择最佳姿势。
- Paper:DiffDock-PP: Rigid Protein-Protein Docking with Diffusion Models
- Github:https://github.com/ketatam/DiffDock-PP
算法整体思路与 DiffDock 接近,Paper:DiffDock: Diffusion Steps, Twists, and Turns for Molecular Docking
Conda 环境配置:
conda create -n diffdock_pp python=3.10.8
conda activate diffdock_ppconda install pytorch=1.13.0 pytorch-cuda=11.6 -c pytorch -c nvidia
验证 PyTorch 是否安装成功:
import torch
print(torch.__version__) # 1.13.0
print(torch.cuda.is_available()) # True
安装其他包:
pip install --no-cache-dir torch-scatter==2.0.9 torch-sparse==0.6.15 torch-cluster torch-spline-conv torch-geometric -f https://data.pyg.org/whl/torch-1.13.0+cu116.htmlpip install numpy dill tqdm pyyaml pandas biopandas scikit-learn biopython e3nn wandb tensorboard tensorboardX matplotlib
下载数据,具体数据路径 datasets/DIPS/pairs_pruned,即:
bypy downfile /psp_data/diffdock_pp/DIPS.zip DIPS.zip
具体使用位于 src/inference.sh,参考 dips_esm_inference.yaml:
data:dataset: dipsdata_file: datasets/DIPS/data_file_100_test.csvdata_path: datasets/DIPS/pairs_pruned
...
其中,数据文件 data_file_100_test.csv,即:
path,split
eb/1ebo.pdb2_1.dill,test
dm/3dmp.pdb3_2.dill,test
kq/1kq1.pdb1_1.dill,test
b2/2b24.pdb1_8.dill,test
cf/3cf0.pdb1_6.dill,test
...
还需要下载 ESM 的 650M 模型,位于 torchhub/checkpoints,否则下载很慢,即:
esm2_t33_650M_UR50D-contact-regression.pt
esm2_t33_650M_UR50D.pt
其中数据类型是 dill 类型,PDB 转换成 dill 类型,参考 https://github.com/octavian-ganea/equidock_public#dips-data
运行推理脚本:
sh src/inference.sh
其中,在运行时,在 src/geom_utils/so3.py 中,需要预处理 npy ,耗时较长,即:
.so3_cdf_vals2.npy
.so3_exp_score_norms2.npy
.so3_omegas_array2.npy
.so3_score_norms2.npy
运行日志:
SCORE_MODEL_PATH: checkpoints/large_model_dips/fold_0/
CONFIDENCE_MODEL_PATH: checkpoints/large_model_dips/fold_0/
SAVE_PATH: ckpts/test_large_model_dips
09:50:16 Starting Inference
data loading: 100%|█| 100/100 [00:00<00:00, 561486
09:50:18 Computing ESM embeddings
Using cache found in torchhub/facebookresearch_esm_main
ESM: 100%|██████████| 4/4 [00:10<00:00, 2.70s/it]
ESM: 100%|██████████| 4/4 [00:06<00:00, 1.61s/it]
09:50:50 finished tokenizing residues with ESM
09:50:50 finished tokenizing all inputs
09:50:50 100 entries loaded
09:50:50 finished loading raw data
09:50:50 running inference
09:50:50 finished creating data splits
miniconda3/envs/diffdock_pp/lib/python3.10/site-packages/torch/jit/_check.py:181: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.warnings.warn("The TorchScript type system doesn't support "
09:50:52 loaded model with kwargs:
checkpoint checkpoints/large_model_dips/fold_0/model_best_338669_140_31.084_30.347.pth
09:50:52 loaded checkpoint from checkpoints/large_model_dips/fold_0/model_best_338669_140_31.084_30.347.pth
09:50:53 loaded model with kwargs:
checkpoint checkpoints/confidence_model_dips/fold_0/model_best_0_6_0.241_0.887.pth
09:50:53 loaded checkpoint from checkpoints/confidence_model_dips/fold_0/model_best_0_6_0.241_0.887.pth
09:50:53 finished loading model
args.temp_sampling: 2.4390%| | 0/100 [00:00<?, ?it/s]09:53:42 Completed 0 out of 40 steps
09:53:44 Completed 1 out of 40 steps
09:53:46 Completed 2 out of 40 steps
09:53:48 Completed 3 out of 40 steps
...
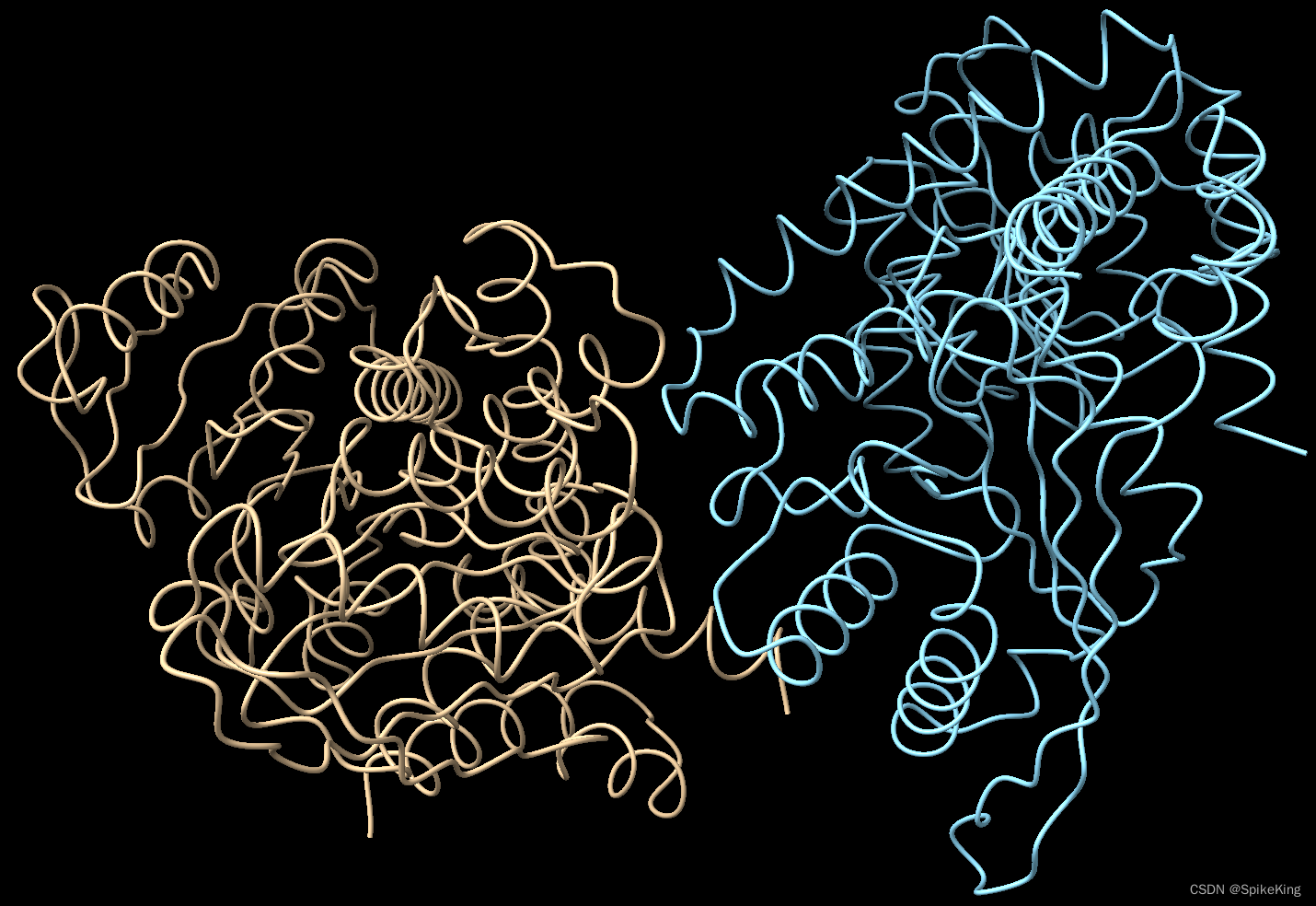
预测结果,1B26 Receptor 和 Ligand,即:
NVCC Bug,参考 GitHub - Failed building wheel for torch-cluster:
In file included from csrc/cuda/graclus_cuda.cu:3:0:diffdock_pp/lib/python3.10/site-packages/torch/include/ATen/cuda/CUDAContext.h:10:10: fatal error: cusolverDn.h: No such file or directory#include <cusolverDn.h>^~~~~~~~~~~~~~compilation terminated.error: command 'diffdock_pp/bin/nvcc' failed with exit code 1[end of output]note: This error originates from a subprocess, and is likely not a problem with pip.ERROR: Failed building wheel for torch-clusterRunning setup.py clean for torch-cluster
Failed to build torch-cluster
ERROR: Could not build wheels for torch-cluster, which is required to install pyproject.toml-based projects
解决方案,参考 [BUG] fatal error: cusolverDn.h: No such file or directory:
export PATH=/usr/local/cuda/bin:$PATH
这篇关于PSP - 蛋白质与蛋白质的扩散对接 DiffDock-PP 算法的文章就介绍到这儿,希望我们推荐的文章对编程师们有所帮助!







