本文主要是介绍unet脑肿瘤分割完整代码,希望对大家解决编程问题提供一定的参考价值,需要的开发者们随着小编来一起学习吧!
U-net脑肿瘤分割完整代码
- 代码目录
- 数据集
- 网络
- 训练
- 测试
代码目录

数据集
https://www.kaggle.com/datasets/mateuszbuda/lgg-mri-segmentation
dataset.py
在这里插入代码片import os
import numpy as np
import glob
from PIL import Image
import cv2
import torchvision
from torch.utils.data import Dataset, DataLoader
from torchvision import transforms
import torch
import matplotlib.pyplot as pltkaggle_3m='./kaggle_3m/'
dirs=glob.glob(kaggle_3m+'*')
#print(dirs)
#os.listdir('./kaggle_3m\\TCGA_HT_A61B_19991127')
data_img=[]
data_label=[]
for subdir in dirs:dirname=subdir.split('\\')[-1]for filename in os.listdir(subdir):img_path=subdir+'/'+filename #图片的绝对路径if 'mask' in img_path:data_label.append(img_path)else:data_img.append(img_path)
#data_img[:5] #前几张图 和标签是否对应
#data_label[:5]
data_imgx=[]
for i in range(len(data_label)):#图片和标签对应img_mask=data_label[i]img=img_mask[:-9]+'.tif'data_imgx.append(img)
#data_imgx
data_newimg=[]
data_newlabel=[]
for i in data_label:#获取只有病灶的数据value=np.max(cv2.imread(i))try:if value>0:data_newlabel.append(i)i_img=i[:-9]+'.tif'data_newimg.append(i_img)except:pass
#查看结果
#data_newimg[:5]
#data_newlabel[:5]
im=data_newimg[20]
im=Image.open(im)
#im.show(im)
im=data_newlabel[20]
im=Image.open(im)
#im.show(im)
#print("可用数据:")
#print(len(data_newlabel))
#print(len(data_newimg))
#数据转换
train_transformer=transforms.Compose([transforms.Resize((256,256)),transforms.ToTensor(),
])
test_transformer=transforms.Compose([transforms.Resize((256,256)),transforms.ToTensor()
])
class BrainMRIdataset(Dataset):def __init__(self, img, mask, transformer):self.img = imgself.mask = maskself.transformer = transformerdef __getitem__(self, index):img = self.img[index]mask = self.mask[index]img_open = Image.open(img)img_tensor = self.transformer(img_open)mask_open = Image.open(mask)mask_tensor = self.transformer(mask_open)mask_tensor = torch.squeeze(mask_tensor).type(torch.long)return img_tensor, mask_tensordef __len__(self):return len(self.img)
s=1000#划分训练集和测试集
train_img=data_newimg[:s]
train_label=data_newlabel[:s]
test_img=data_newimg[s:]
test_label=data_newlabel[s:]
#加载数据
train_data=BrainMRIdataset(train_img,train_label,train_transformer)
test_data=BrainMRIdataset(test_img,test_label,test_transformer)dl_train=DataLoader(train_data,batch_size=4,shuffle=True)
dl_test=DataLoader(test_data,batch_size=4,shuffle=True)img,label=next(iter(dl_train))
plt.figure(figsize=(12,8))
for i,(img,label) in enumerate(zip(img[:4],label[:4])):img=img.permute(1,2,0).numpy()label=label.numpy()plt.subplot(2,4,i+1)plt.imshow(img)plt.subplot(2,4,i+5)plt.imshow(label)
网络
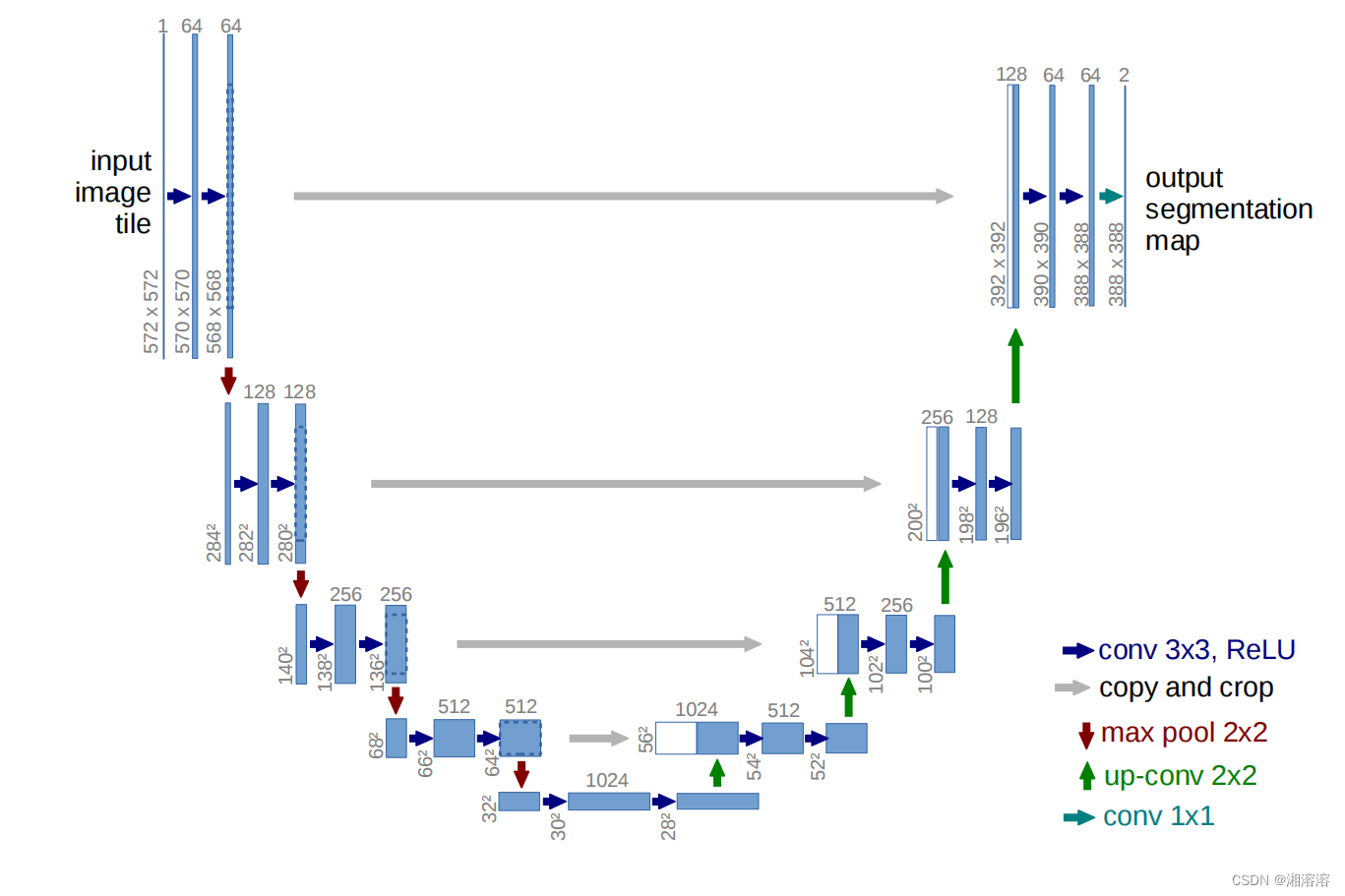
model.py
import torch
import torch.nn as nnclass Downsample(nn.Module):def __init__(self, in_channels, out_channels):super(Downsample, self).__init__()self.conv_relu = nn.Sequential(nn.Conv2d(in_channels, out_channels,kernel_size=3, padding=1),nn.ReLU(inplace=True),nn.Conv2d(out_channels, out_channels,kernel_size=3, padding=1),nn.ReLU(inplace=True))self.pool = nn.MaxPool2d(kernel_size=2)def forward(self, x, is_pool=True):if is_pool:x = self.pool(x)x = self.conv_relu(x)return xclass Upsample(nn.Module):def __init__(self, channels):super(Upsample, self).__init__()self.conv_relu = nn.Sequential(nn.Conv2d(2 * channels, channels,kernel_size=3, padding=1),nn.ReLU(inplace=True),nn.Conv2d(channels, channels,kernel_size=3, padding=1),nn.ReLU(inplace=True))self.upconv_relu = nn.Sequential(nn.ConvTranspose2d(channels,channels // 2,kernel_size=3,stride=2,padding=1,output_padding=1),nn.ReLU(inplace=True))def forward(self, x):x = self.conv_relu(x)x = self.upconv_relu(x)return xclass Net(nn.Module):def __init__(self):super(Net, self).__init__()self.down1 = Downsample(3, 64)self.down2 = Downsample(64, 128)self.down3 = Downsample(128, 256)self.down4 = Downsample(256, 512)self.down5 = Downsample(512, 1024)self.up = nn.Sequential(nn.ConvTranspose2d(1024,512,kernel_size=3,stride=2,padding=1,output_padding=1),nn.ReLU(inplace=True))self.up1 = Upsample(512)self.up2 = Upsample(256)self.up3 = Upsample(128)self.conv_2 = Downsample(128, 64)self.last = nn.Conv2d(64, 2, kernel_size=1)def forward(self, x):x1 = self.down1(x, is_pool=False)x2 = self.down2(x1)x3 = self.down3(x2)x4 = self.down4(x3)x5 = self.down5(x4)x5 = self.up(x5)x5 = torch.cat([x4, x5], dim=1) # 32*32*1024x5 = self.up1(x5) # 64*64*256)x5 = torch.cat([x3, x5], dim=1) # 64*64*512x5 = self.up2(x5) # 128*128*128x5 = torch.cat([x2, x5], dim=1) # 128*128*256x5 = self.up3(x5) # 256*256*64x5 = torch.cat([x1, x5], dim=1) # 256*256*128x5 = self.conv_2(x5, is_pool=False) # 256*256*64x5 = self.last(x5) # 256*256*3return x5if __name__ == '__main__':x = torch.rand([8, 3, 256, 256])model = Net()y = model(x)
训练
train.py
import torch as t
import torch.nn as nn
from tqdm import tqdm #进度条
import model
from dataset import *device = t.device("cuda") if t.cuda.is_available() else t.device("cpu")train_data=BrainMRIdataset(train_img,train_label,train_transformer)
test_data=BrainMRIdataset(test_img,test_label,test_transformer)dl_train=DataLoader(train_data,batch_size=4,shuffle=True)
dl_test=DataLoader(test_data,batch_size=4,shuffle=True)model = model.Net()
img,label=next(iter(dl_train))
model=model.to('cuda')
img=img.to('cuda')
pred=model(img)
label=label.to('cuda')
loss_fn=nn.CrossEntropyLoss()#交叉熵损失函数
loss_fn(pred,label)
optimizer=torch.optim.Adam(model.parameters(),lr=0.0001)
def train_epoch(epoch, model, trainloader, testloader):correct = 0total = 0running_loss = 0epoch_iou = [] #交并比net=model.train()for x, y in tqdm(testloader):x, y = x.to('cuda'), y.to('cuda')y_pred = model(x)loss = loss_fn(y_pred, y)optimizer.zero_grad()loss.backward()optimizer.step()with torch.no_grad():y_pred = torch.argmax(y_pred, dim=1)correct += (y_pred == y).sum().item()total += y.size(0)running_loss += loss.item()intersection = torch.logical_and(y, y_pred)union = torch.logical_or(y, y_pred)batch_iou = torch.sum(intersection) / torch.sum(union)epoch_iou.append(batch_iou.item())epoch_loss = running_loss / len(trainloader.dataset)epoch_acc = correct / (total * 256 * 256)test_correct = 0test_total = 0test_running_loss = 0epoch_test_iou = []t.save(net.state_dict(), './Results/weights/unet_weight/{}.pth'.format(epoch))model.eval()with torch.no_grad():for x, y in tqdm(testloader):x, y = x.to('cuda'), y.to('cuda')y_pred = model(x)loss = loss_fn(y_pred, y)y_pred = torch.argmax(y_pred, dim=1)test_correct += (y_pred == y).sum().item()test_total += y.size(0)test_running_loss += loss.item()intersection = torch.logical_and(y, y_pred)#预测值和真实值之间的交集union = torch.logical_or(y, y_pred)#预测值和真实值之间的并集batch_iou = torch.sum(intersection) / torch.sum(union)epoch_test_iou.append(batch_iou.item())epoch_test_loss = test_running_loss / len(testloader.dataset)epoch_test_acc = test_correct / (test_total * 256 * 256)#预测正确的值除以总共的像素点print('epoch: ', epoch,'loss: ', round(epoch_loss, 3),'accuracy:', round(epoch_acc, 3),'IOU:', round(np.mean(epoch_iou), 3),'test_loss: ', round(epoch_test_loss, 3),'test_accuracy:', round(epoch_test_acc, 3),'test_iou:', round(np.mean(epoch_test_iou), 3))return epoch_loss, epoch_acc, epoch_test_loss, epoch_test_accif __name__ == "__main__":epochs=20for epoch in range(epochs):train_epoch(epoch,model,dl_train,dl_test)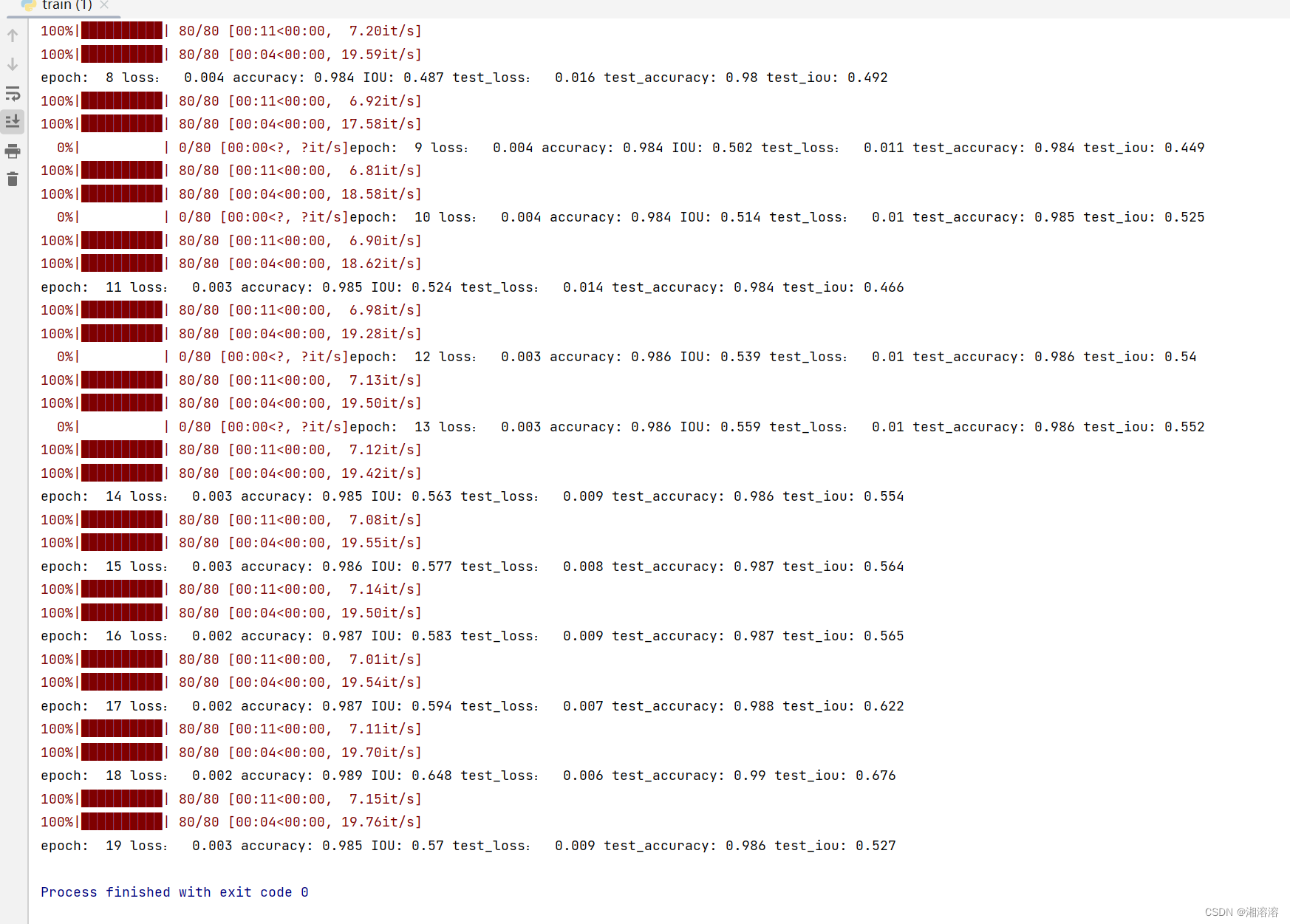
只跑了20个epoch
测试
test.py
import torch as t
import torch.nn as nn
import model
from dataset import *
import matplotlib.pyplot as pltdevice = t.device("cuda") if t.cuda.is_available() else t.device("cpu")train_data=BrainMRIdataset(train_img,train_label,train_transformer)
test_data=BrainMRIdataset(test_img,test_label,test_transformer)dl_train=DataLoader(train_data,batch_size=4,shuffle=True)
dl_test=DataLoader(test_data,batch_size=4,shuffle=True)model = model.Net()
img,label=next(iter(dl_train))
model=model.to('cuda')
img=img.to('cuda')
pred=model(img)
label=label.to('cuda')
loss_fn=nn.CrossEntropyLoss()
loss_fn(pred,label)
optimizer=torch.optim.Adam(model.parameters(),lr=0.0001)
def test():image, mask = next(iter(dl_test))image=image.to('cuda')net = model.eval()net.to(device)net.load_state_dict(t.load("./Results/weights/unet_weight/18.pth"))pred_mask = model(image)pred_mask=pred_maskmask=torch.squeeze(mask)pred_mask=pred_mask.cpu()num=4plt.figure(figsize=(10, 10))for i in range(num):plt.subplot(num, 4, i*num+1)plt.imshow(image[i].permute(1,2,0).cpu().numpy())plt.subplot(num, 4, i*num+2)plt.imshow(mask[i].cpu().numpy(),cmap='gray')#标签plt.subplot(num, 4, i*num+3)plt.imshow(torch.argmax(pred_mask[i].permute(1,2,0), axis=-1).detach().numpy(),cmap='gray')#预测plt.show()if __name__ == "__main__":test()模型分割效果
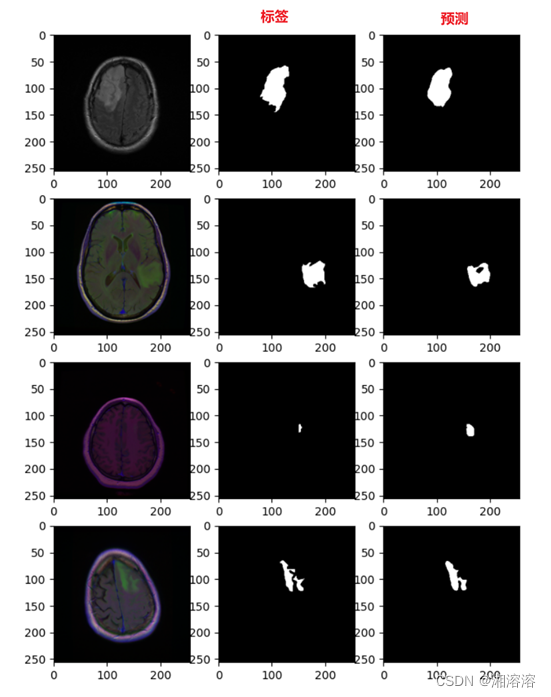
这篇关于unet脑肿瘤分割完整代码的文章就介绍到这儿,希望我们推荐的文章对编程师们有所帮助!





