本文主要是介绍PSP - 替换 MSA (多序列比对) 文件的 Target 序列,希望对大家解决编程问题提供一定的参考价值,需要的开发者们随着小编来一起学习吧!
欢迎关注我的CSDN:https://spike.blog.csdn.net/
本文地址:https://blog.csdn.net/caroline_wendy/article/details/131898038
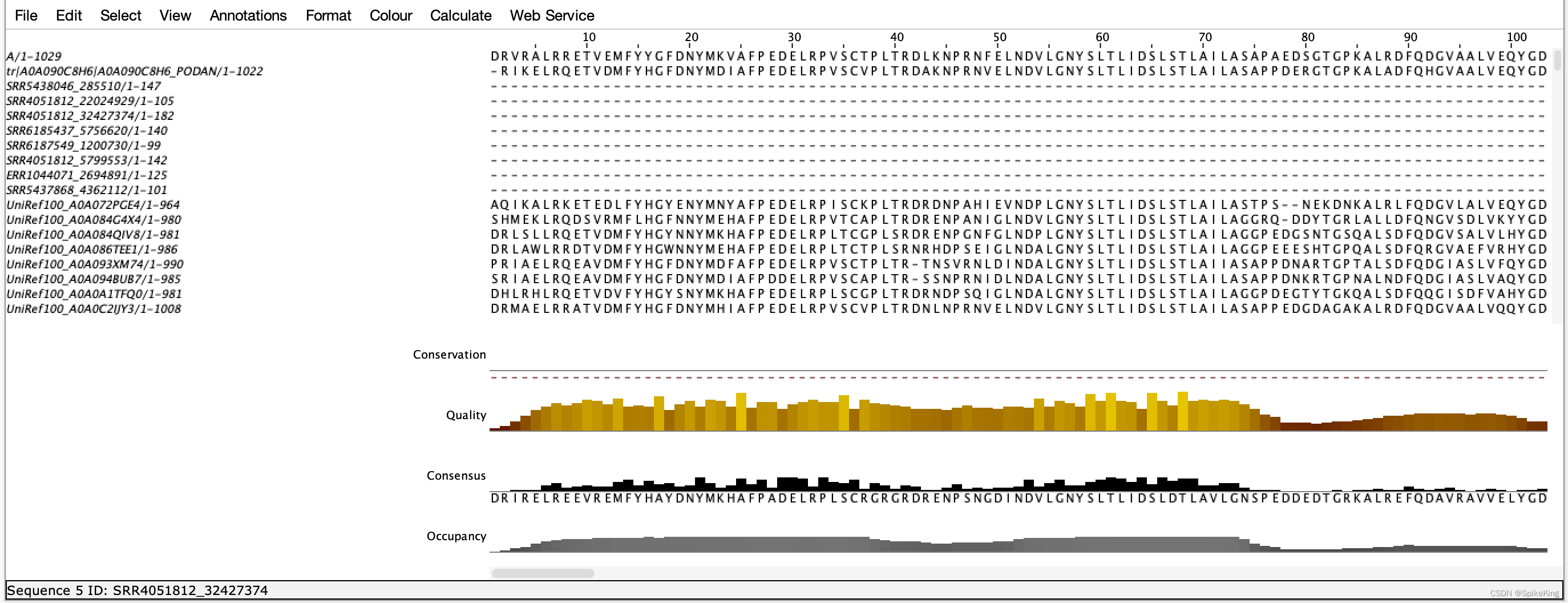
在 MSA 文件中,通过处理 Target 序列,影响 MSA 的搜索结果与比对结果,但是在预测的过程中,需要替换为最初的 Target 序列,同时,保持序列长度一致。
常见场景是去除 IDRs 无序区域,进行 MSA 搜索,再替换目标序列,进行蛋白质结构预测。参考:
- 合并 AlphaFold2 MSA 搜索的全部文件 (a3m or sto)
- MetaPredict 预测蛋白质序列的内源性无序区域 (IDRs)
即:
- 读取 MSA 文件。
- 替换 MSA 中 Target 序列为目标序列 (FASTA文件)。
- 再存储至原 MSA 文件。
源码如下:
#!/usr/bin/env python
# -- coding: utf-8 --
"""
Copyright (c) 2022. All rights reserved.
Created by C. L. Wang on 2023/7/24
"""
import argparse
import os
import sys
from pathlib import Pathp = os.path.dirname(os.path.dirname(os.path.abspath(__file__)))
if p not in sys.path:sys.path.append(p)from myutils.project_utils import read_file, write_list_to_file, create_empty_file
from protein_utils.seq_utils import get_seq_from_fastaclass MsaReplaceTarget(object):"""替换 MSA 的 Target 序列,即第 1 条序列"""def __init__(self):pass@staticmethoddef process(fasta_path, mas_path):assert os.path.isfile(fasta_path) and os.path.isfile(mas_path)t_seq_list, t_desc_list = get_seq_from_fasta(fasta_path)t_seq, t_desc = t_seq_list[0], t_desc_list[0]print(f"[Info] t_desc: {t_desc}")print(f"[Info] t_seq: {t_seq}")data_lines = read_file(mas_path)data_lines[1] = t_seq # 只替换seqassert len(data_lines[1]) == len(data_lines[3]) # 保证序列长度一致create_empty_file(mas_path)write_list_to_file(mas_path, data_lines)print("[Info] 处理完成! ")def main():parser = argparse.ArgumentParser()parser.add_argument("-f","--fasta-path",help="the fasta path of target.",type=Path,required=True,)parser.add_argument("-m","--msa-path",help="the msa file",type=Path,required=True)args = parser.parse_args()fasta_path = str(args.fasta_path)msa_path = str(args.msa_path)assert os.path.isfile(fasta_path) and os.path.isfile(msa_path)cms = MsaReplaceTarget()cms.process(fasta_path, msa_path)if __name__ == '__main__':main()
替换之前的 MSA 序列:
>A
XXVRALRRETVEMFYYGFDNYMKVAFPEDELRPVSCTPLTRDLKNPRNFELNDVLGNYSLTLIDSLSTLAILASAPAEDSGTGPKALRDFQDGVAALVEQYGDGRPGPSGVGRRARGFDLDSKVQVFETVIRGVGGLLSAHLFAIGALPITGYQPLRQEDDLFNPPPIPWPNGFTYDGQLLRLALDLAQRLLPAFYTKTGLPYPRVNLRHGIPFYVNSPLHEDPXXXXXXXGPPEITETCSAGAGSLVLEFTVLSRLTGDPRFEQAAKRAFWAVWYRKSQIGLIGAGVDAEQGHWIGTYSVIGAGADSFFEYALKSHILLSGHALPNQTHPSPLHKDVNWMDPNTLFEPLSDAENSAESFLEAWHHAHAAIKRHLYSEREHPHYDNVNLWTGSLVSHWVDSLGAYYSGLLVLAGEVDEAIETNLLYAAIWTRYAALPERWSLREKTVEGGLGWWPLRPEFIESTYHLYRATKDPWYLYVGEMVLRDITRRCWTPCGWAGLQNVLSGEKSDRMESFFLGETTKYMYLLFDDDHPLNKLDASFVFTTEGHPLILPXXXXXXXXXXXXXXXXXXLTVYQGEGFTNSCPPRPSITPLSGSVIAARDDIYHPARMVDLHLLTTSKHALDGGQMSGQHMAKSNYTLYPWTLPPELLPSNGTCAKVYQPHEVTLEFASNTQQVLGGSAFNFMLSGQNLERLSTDRIRVLSLSGLKITLQLVEEGEREWRVTKLNGIPLGRDEYVVINRAILGDVSDPRFNLVRDPVIAKLQQLHQVNLLXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXSALLPDLSSFVKSLFARLSNLXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXPVPESLFPWKTIYAAGEACAGPLPDSAPRENQVILIRRGGCSFSDKLANIPAFTPSEESLQLVVVVSDDEHEGQSGLVRPLLDEIQHTPGGMPRRHPIAMVMVGGGETVYQQLSVASAIGIQRRYYIESSGVKVKNIIVDXXXXXXXX
>tr|A0A090C8H6|A0A090C8H6_PODAN Putative Glycoside Hydrolase Family 47 OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) PE=4 SV=1
-RIKELRQETVDMFYHGFDNYMDIAFPEDELRPVSCVPLTRDAKNPRNVELNDVLGNYSLTLIDSLSTLAILASAPPDERGTGPKALADFQHGVAALVEQYGDGSPGPSGVGQRGRGFDVDSKVQVFETVIRGLGGLLSAHLFAVGALPITGYKPRHIEDDPLYSQPIVWPNGFKYDGQILRLALDLGQRLLPAFYTKTGMPYPRVNLRHGIPFYTNSPMHENAPM-NPPEGPLEITETCSAGAGSLVLEFTVLSRLTGDPRFEQLAKRAFWAVWYRKSQIGLIGAGVDAEQGHWIGAYAVIGAGADSFFEYALKSHILLSGHEPPNRTAPARKHRSDNWLDPNALFPPLNDAENSADSFLEAWHLAHAAIKRHLYNEKDHPHYDNVNLWTGSLVSNWVDSLGAYYSGLLVLAGEVEEAIETNLLYTAIWTRYAALPERYSLRDKTVEGGLGWWPLRPEFIESTYHIYRATKDPWYLYVGEMVLRDITRRCWTPCGWAGLQNVLDGEKSDRMESFFLGETAKYMYLLFDDEHPLNSLDAPYVFTTEGHPLIIPKAPPKDGPRRR-RSPRKYLTVYPNEEYTNTCPPRPQTTPLSGSVVAARDDIYHAARLLDLHQLSPTSAAIDAGQMSGQHMARSNYTLYPWTLPAELMPDNGICAKLYQPEEVTLEFASNAQQAVGGSSFNFLLGSQNLERLSADRIRVSSLSGLKMSMRLEDTGEREWRVSKVNGVLLGKDESIIFDRAILGEIQDPRFSLIKDPVLAKLQQLHQINLLDDEPAASDDGRKAGQQPLSQTEDTHEEELDADLPPVASPRVSVPAFGSMVKALFNQIAASLDLQLPDATSIPGLRSSTPKKAPINRVTPAAPLPAHIIPPRAPRIPEFGPVPIEHFPWSTIYAAGTACDAVLPDSAPRDHQVIVIRRGGCNFSTKLANIPAFSPSFRSLQLVVVVSDDHLREQAGLIRPLLDEVQVTPAGFARRHPIPMVMVGGGDVGYEQLGAAKRMGLARRWFVESSGFRVRNVIVDEGDN----
调用脚本,替换之后的 MSA 序列:
>A
DRVRALRRETVEMFYYGFDNYMKVAFPEDELRPVSCTPLTRDLKNPRNFELNDVLGNYSLTLIDSLSTLAILASAPAEDSGTGPKALRDFQDGVAALVEQYGDGRPGPSGVGRRARGFDLDSKVQVFETVIRGVGGLLSAHLFAIGALPITGYQPLRQEDDLFNPPPIPWPNGFTYDGQLLRLALDLAQRLLPAFYTKTGLPYPRVNLRHGIPFYVNSPLHEDPPAKGTTEGPPEITETCSAGAGSLVLEFTVLSRLTGDPRFEQAAKRAFWAVWYRKSQIGLIGAGVDAEQGHWIGTYSVIGAGADSFFEYALKSHILLSGHALPNQTHPSPLHKDVNWMDPNTLFEPLSDAENSAESFLEAWHHAHAAIKRHLYSEREHPHYDNVNLWTGSLVSHWVDSLGAYYSGLLVLAGEVDEAIETNLLYAAIWTRYAALPERWSLREKTVEGGLGWWPLRPEFIESTYHLYRATKDPWYLYVGEMVLRDITRRCWTPCGWAGLQNVLSGEKSDRMESFFLGETTKYMYLLFDDDHPLNKLDASFVFTTEGHPLILPKPKSARRSRNSPRSSQKALTVYQGEGFTNSCPPRPSITPLSGSVIAARDDIYHPARMVDLHLLTTSKHALDGGQMSGQHMAKSNYTLYPWTLPPELLPSNGTCAKVYQPHEVTLEFASNTQQVLGGSAFNFMLSGQNLERLSTDRIRVLSLSGLKITLQLVEEGEREWRVTKLNGIPLGRDEYVVINRAILGDVSDPRFNLVRDPVIAKLQQLHQVNLLDDTTTEEHPDNLDTLDTASAIDLPQDQSSDSEVPDPANLSALLPDLSSFVKSLFARLSNLTSPSPDPSSNLPLNVVINQTAILPTGIGAAPLPPAASNSPSGAPIPVFGPVPESLFPWKTIYAAGEACAGPLPDSAPRENQVILIRRGGCSFSDKLANIPAFTPSEESLQLVVVVSDDEHEGQSGLVRPLLDEIQHTPGGMPRRHPIAMVMVGGGETVYQQLSVASAIGIQRRYYIESSGVKVKNIIVDDGDGGVDG
>tr|A0A090C8H6|A0A090C8H6_PODAN Putative Glycoside Hydrolase Family 47 OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) PE=4 SV=1
-RIKELRQETVDMFYHGFDNYMDIAFPEDELRPVSCVPLTRDAKNPRNVELNDVLGNYSLTLIDSLSTLAILASAPPDERGTGPKALADFQHGVAALVEQYGDGSPGPSGVGQRGRGFDVDSKVQVFETVIRGLGGLLSAHLFAVGALPITGYKPRHIEDDPLYSQPIVWPNGFKYDGQILRLALDLGQRLLPAFYTKTGMPYPRVNLRHGIPFYTNSPMHENAPM-NPPEGPLEITETCSAGAGSLVLEFTVLSRLTGDPRFEQLAKRAFWAVWYRKSQIGLIGAGVDAEQGHWIGAYAVIGAGADSFFEYALKSHILLSGHEPPNRTAPARKHRSDNWLDPNALFPPLNDAENSADSFLEAWHLAHAAIKRHLYNEKDHPHYDNVNLWTGSLVSNWVDSLGAYYSGLLVLAGEVEEAIETNLLYTAIWTRYAALPERYSLRDKTVEGGLGWWPLRPEFIESTYHIYRATKDPWYLYVGEMVLRDITRRCWTPCGWAGLQNVLDGEKSDRMESFFLGETAKYMYLLFDDEHPLNSLDAPYVFTTEGHPLIIPKAPPKDGPRRR-RSPRKYLTVYPNEEYTNTCPPRPQTTPLSGSVVAARDDIYHAARLLDLHQLSPTSAAIDAGQMSGQHMARSNYTLYPWTLPAELMPDNGICAKLYQPEEVTLEFASNAQQAVGGSSFNFLLGSQNLERLSADRIRVSSLSGLKMSMRLEDTGEREWRVSKVNGVLLGKDESIIFDRAILGEIQDPRFSLIKDPVLAKLQQLHQINLLDDEPAASDDGRKAGQQPLSQTEDTHEEELDADLPPVASPRVSVPAFGSMVKALFNQIAASLDLQLPDATSIPGLRSSTPKKAPINRVTPAAPLPAHIIPPRAPRIPEFGPVPIEHFPWSTIYAAGTACDAVLPDSAPRDHQVIVIRRGGCNFSTKLANIPAFSPSFRSLQLVVVVSDDHLREQAGLIRPLLDEVQVTPAGFARRHPIPMVMVGGGDVGYEQLGAAKRMGLARRWFVESSGFRVRNVIVDEGDN----
这篇关于PSP - 替换 MSA (多序列比对) 文件的 Target 序列的文章就介绍到这儿,希望我们推荐的文章对编程师们有所帮助!




