本文主要是介绍Epicypher—CUTANA™ ChIC/CUTRUN Kit,希望对大家解决编程问题提供一定的参考价值,需要的开发者们随着小编来一起学习吧!

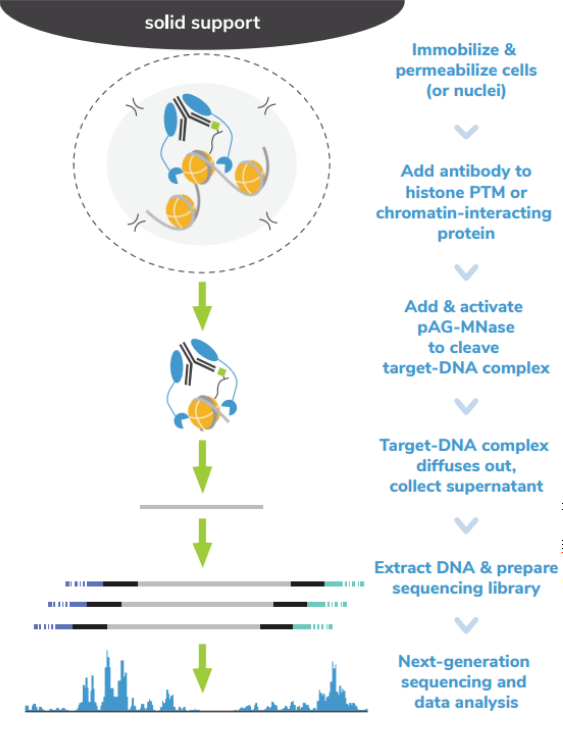
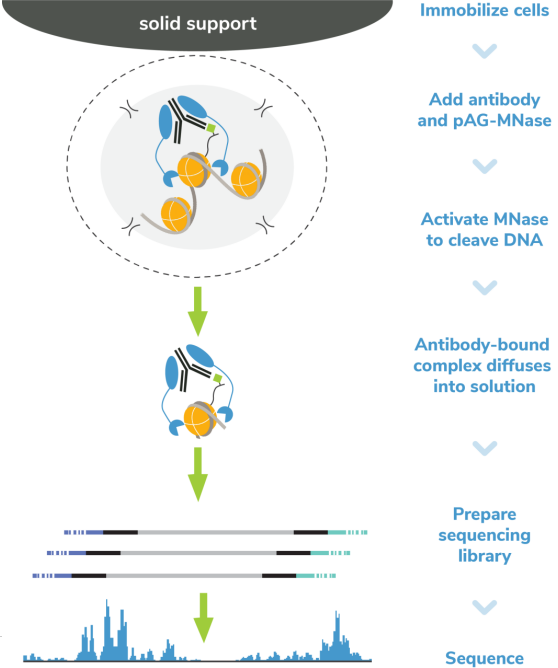
核酸酶靶向切割和释放 (CUT&RUN)技术是由Steven henikoff博士团队开发的一种染色质图谱分析方法,基于Ulrich Laemmli博士的染色质免疫切割技术 (ChIC),融合蛋白A与微球菌核酸酶 (pA-MNase),选择性原位切割与抗体结合的染色质。在CUT&RUN中,细胞或细胞核固定化在固相载体上,从溶液中分离出pAG-MNase裂解的DNA片段。该方法与二代测序(NGS)兼容,可提供高质量的组蛋白翻译后修饰(PTMs)和染色质相关蛋白(如转录因子;Figure 1)。
ChIP-seq是组蛋白PTMs和染色质相关蛋白全基因组定位的主要方法。在这种方法中,染色质通过超声或酶消化破碎,然后免疫沉淀目标特异性片段。尽管进行优化,但ChIP-seq需要大量细胞(通常为105 - 106个细胞)而且需要深度测序input 染色质与免疫沉淀物质(通常为 >3000万 reads/次)来从背景中解析信号。
ChIC和CUT&RUN通过将基因组片段靶向释放到溶液中,彻底改变了染色质调控。通过这一创新,背景显著减少,允许使用少量细胞且每个反应仅需300 - 800万reads/次对组蛋白PTMs和染色质相关蛋白进行高分辨率基因组图谱的绘制(Figure 2)。简化的工作流程和节省的成本使ChIC/CUT&RUN适用于高通量研究表观遗传生物学。
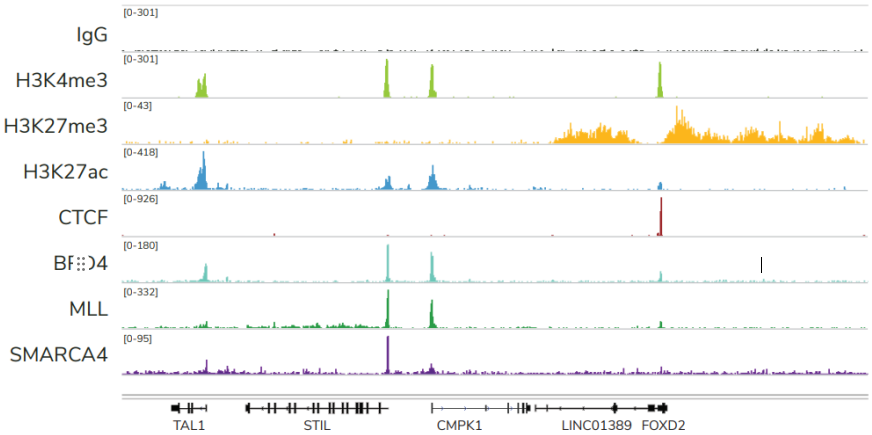
FIGURE 2 Representative genome browser tracks show CUTANA™ CUT&RUN results using 500,000 K562 cells. Clear peaks with the expected distribution profile are observed using 3-8 million sequencing reads per reaction for a variety of epigenetic targets, including histone PTMs (H3K4me3, H3K27me3, H3K27ac), transcription factors (CTCF), epigenetic reader proteins (BRD4), writer enzymes (MLL1), and chromatin remodelers (SMARCA4). Rabbit IgG antibody shown as a negative control (top track).
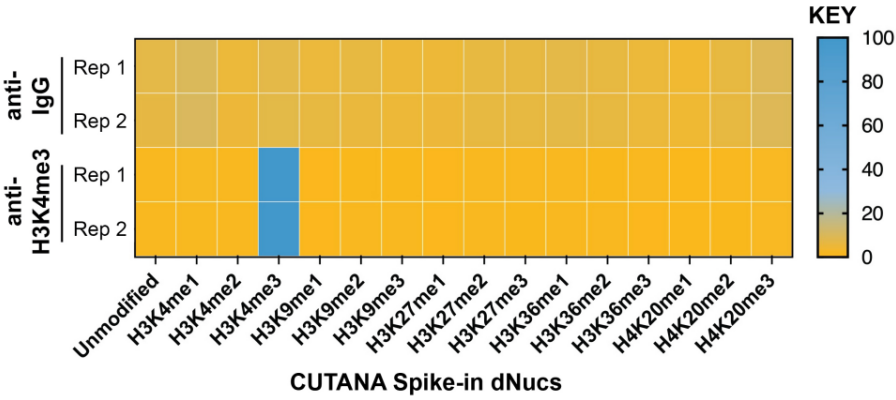
CUTANA™ChIC/CUT&RUN试剂盒包含48个反应的材料,专为多通道移液而设计,以实现CUT&RUN的高通量优势。该试剂盒包括阳性(H3K4me3)和阴性(Rabbit IgG)对照抗体,以及SNAP-CUTANA™ K-MetStat Panel (16个DNA条形码设计核小体携带广泛研究的赖氨酸甲基化PTMs)的分装分量。K-MetStat Panel加入到对照反应中,直接监测实验成功与否并帮助排除故障。此外,在pAG-MNase切割后,将剪切的E. coli DNA添加到所有反应中,以控制文库构建并使NGS标准化。该试剂盒与细胞和细胞核兼容,包括冻存和交联样品(Figure 3)。
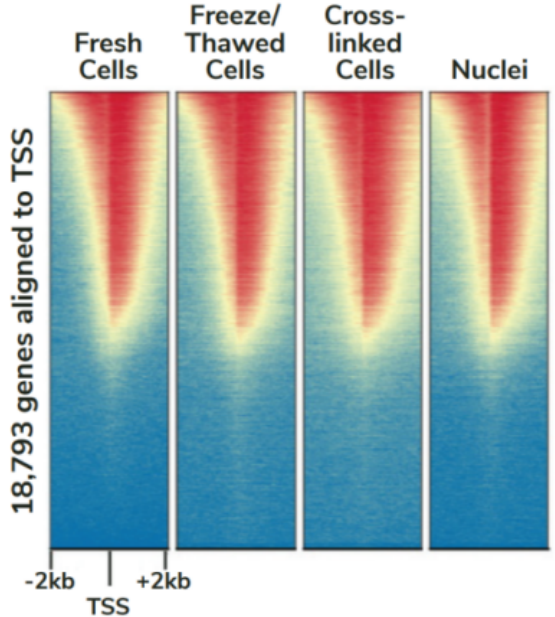
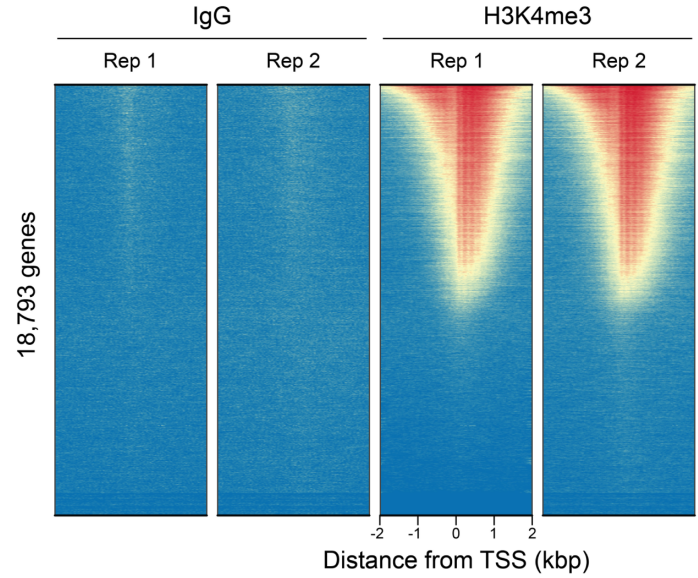
FIGURE 3 Heatmaps show CUT&RUN signal (red) and background (blue) of H3K4me3-enriched regions flanking annotated transcription start sites (TSS, +/- 2 kb). Gene rows are aligned across conditions, showing that genome-wide enrichment is preserved across sample types.
尽管建议从500,000个细胞开始,但只需使用5,000个细胞即可生成可比较的数据(Figure 4)。对照组的加入以及与不同靶类型、样本和低细胞数的兼容性,使该试剂盒成为染色质图谱实验的首选解决方案。
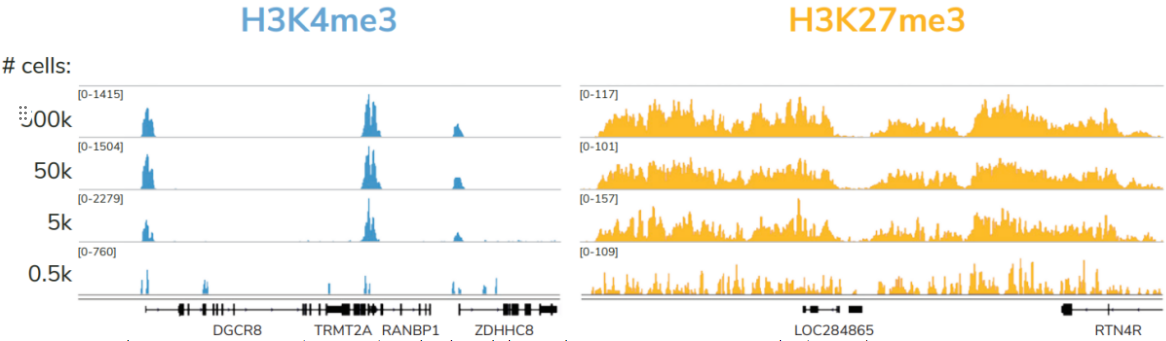
FIGURE 4 Representative genome browser tracks for H3K4me3 (low abundance target) and H3K27me3 (high abundance target) CUT&RUN experiments using decreasing amounts of K562 cells. At 5,000 cells, data quality is largely indistinguishable from standard conditions (500,000 cells).
保存条件
OPEN KIT IMMEDIATELY and store components at room temperature, 4℃, and -20℃ as indicated (see User Manual corresponding to Kit Version 3). Stable for 6 months upon date of receipt.
| Room Temperature (RT) | 4℃ | -20℃ |
| 8-strip Tubes | ConA Beads | 5% Digitonin |
| 0.5 M EDTA | E. coli Spike-in DNA | 1 M Spermidine |
| 100 mM Calcium Chloride | Bead Activation Buffer | SNAP-CUTANA™ K-MetStat Panel |
| SPRIselect Reagent Manufactured by Beckman Coulter Inc. | Pre-Wash Buffer | H3K4me3 Positive Control Antibody |
| 0.1×TE Buffer | Stop Buffer | Rabbit IgG Negative Control Antibody |
| pAG-MNase |
数据示例
|
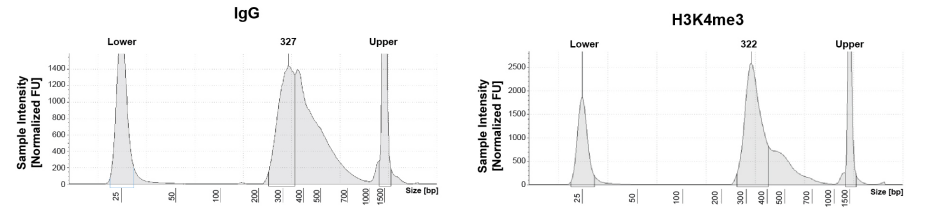
| Figure 1: CUT&RUN DNA fragment size distribution analysis CUT&RUN was performed as described in Figure 5. Library DNA was analyzed by Agilent Tapestation®. This analysis confirmed that mononucleosomes were predominantly enriched in CUT&RUN (~300 bp peaks represent 150 bp nucleosomes + sequencing adapters). |
|
| FIGURE 2 SNAP-CUTANA™ K-MetStat Spikein Controls DNA-barcoded designer nucleosomes (dNucs) representing 16 K-methyl PTMs: mono-, di-, and tri-methylation at H3K4, H3K9, H3K27, H3K36, and H4K20, as well as unmodified control, were spiked into CUT&RUN reactions prior to the addition of antibodies (IgG, H3K4me3). Spike-in barcodes were counted and normalized from raw fastq files using the shell script and analysis sheet available at epicypher.com/19-1002. Barcodes for IgG (top; normalized to total reads) and H3K4me3 (bottom; normalized to on-target) antibodies are shown. The spike-ins confirmed optimal experimental conditions (H3K4me3 antibody specifically recovered the target dNuc, while IgG showed no preferential enrichment). |
|
| Figure 3: CUT&RUN genome-wide heatmaps CUT&RUN was performed as described in Figure 5. Heatmaps show two replicates (“Rep”) of IgG (left) and H3K4me3 (right) kit control antibodies in aligned rows ranked by intensity (top to bottom) and colored such that red indicates high localized enrichment and blue denotes background signal. |
|
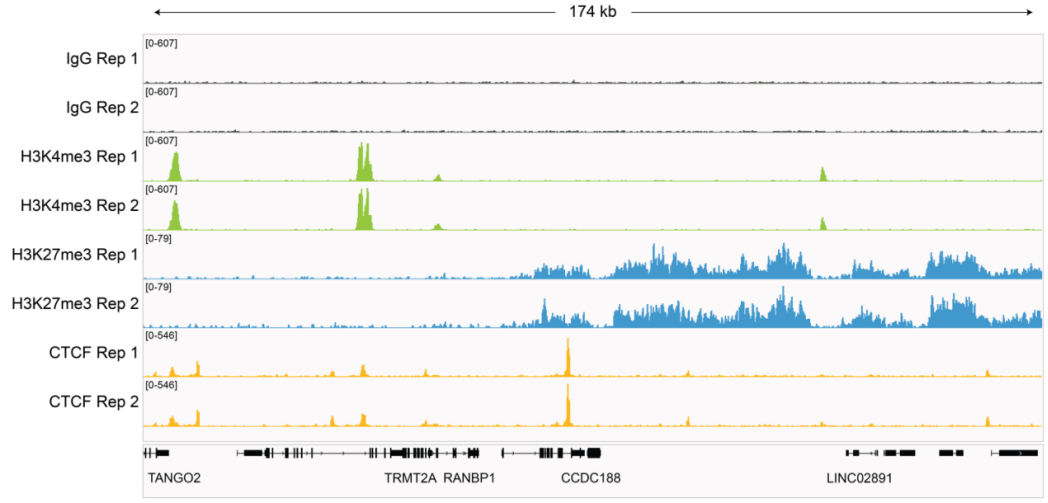
| Figure 4: Representative gene browser tracks CUT&RUN was performed as described in Figure 5. A representative 174 kb window at the TRMT2A gene is shown for two replicates (“Rep”) of IgG and H3K4me3 kit control antibodies. Representative tracks are also shown for antibodies to H3K27me3 and the transcription factor CTCF. The CUT&RUN kit produced the expected genomic distribution for each target. Images were generated using the Integrative Genomics Viewer (IGV, Broad Institute). |
|
| Figure 5: CUT&RUN methods CUT&RUN was performed using the CUTANA™ ChIC/CUT&RUN Kit starting with 500k K562 cells with 0.5 µg of either IgG (EpiCypher 13-0042), H3K4me3 (EpiCypher 13-0041), H3K27me3 (EpiCypher 13-0055), or 0.125 µg of CTCF (EpiCypher 13-2014) antibodies in duplicate. Library preparation was performed with 5 ng of DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ Library Prep Kit (EpiCypher 14-1001/14-1002). Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2x50 bp). Sample sequencing depth was 3.5 million reads (IgG Rep 1), 3.8 million reads (IgG Rep 2), 4.7 million reads (H3K4me3 Rep 1), 6.9 million reads (H3K4me3 Rep 2), 6.6 million reads (H3K27me3 Rep 1), 4.7 million reads (H3K27me3 Rep 2), 3.9 million reads (CTCF Rep 1) and 4.6 million reads (CTCF Rep 2). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and blacklist regions. |
订购详情
| 货号 | 产品名称 | 规格 |
| 14-1048 | CUTANA™ ChIC/CUT&RUN Kit / 欣博盛生物 | 48 Reactions |
这篇关于Epicypher—CUTANA™ ChIC/CUTRUN Kit的文章就介绍到这儿,希望我们推荐的文章对编程师们有所帮助!





![[Golang] go-kit 介绍和使用 (微服务实现工具)](https://img-blog.csdnimg.cn/direct/cdb4a8f75c044dba8c2235333a60117c.png)